| Full name: IGF like family member 2 | Alias Symbol: UNQ645 | ||
| Type: protein-coding gene | Cytoband: 19q13.32 | ||
| Entrez ID: 147920 | HGNC ID: HGNC:32929 | Ensembl Gene: ENSG00000204866 | OMIM ID: 610545 |
| Drug and gene relationship at DGIdb | |||
Expression of IGFL2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | IGFL2 | 147920 | 231148_at | 0.0277 | 0.9771 | |
| GSE26886 | IGFL2 | 147920 | 231148_at | 0.6501 | 0.0332 | |
| GSE45670 | IGFL2 | 147920 | 231148_at | 1.0571 | 0.1175 | |
| GSE53622 | IGFL2 | 147920 | 95013 | 1.6003 | 0.0000 | |
| GSE53624 | IGFL2 | 147920 | 95013 | 2.0202 | 0.0000 | |
| GSE63941 | IGFL2 | 147920 | 231148_at | 0.1949 | 0.4586 | |
| GSE77861 | IGFL2 | 147920 | 231148_at | 0.1571 | 0.5448 | |
| GSE97050 | IGFL2 | 147920 | A_33_P3240602 | -0.0332 | 0.8926 | |
| TCGA | IGFL2 | 147920 | RNAseq | 2.5012 | 0.0000 |
Upregulated datasets: 3; Downregulated datasets: 0.
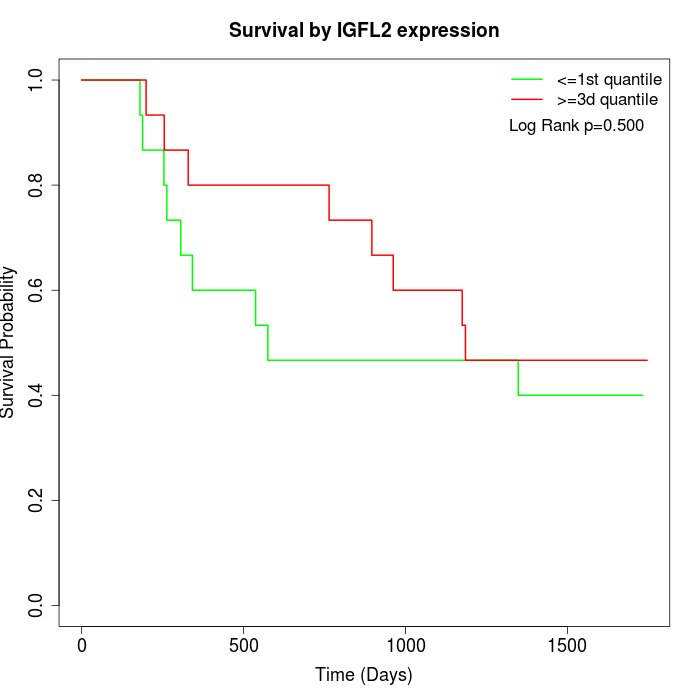
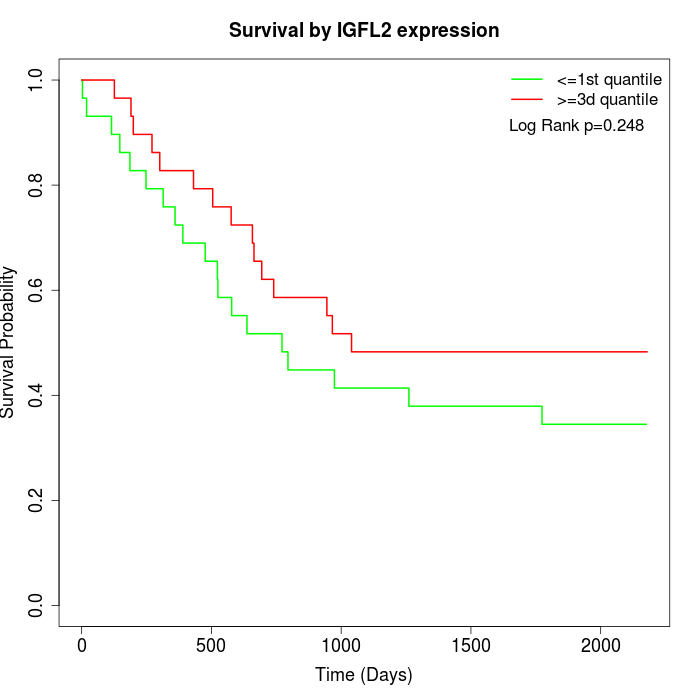
Survival by IGFL2 expression:
Note: Click image to view full size file.
Copy number change of IGFL2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | IGFL2 | 147920 | 4 | 5 | 21 | |
| GSE20123 | IGFL2 | 147920 | 4 | 4 | 22 | |
| GSE43470 | IGFL2 | 147920 | 3 | 11 | 29 | |
| GSE46452 | IGFL2 | 147920 | 45 | 1 | 13 | |
| GSE47630 | IGFL2 | 147920 | 8 | 7 | 25 | |
| GSE54993 | IGFL2 | 147920 | 17 | 4 | 49 | |
| GSE54994 | IGFL2 | 147920 | 5 | 13 | 35 | |
| GSE60625 | IGFL2 | 147920 | 9 | 0 | 2 | |
| GSE74703 | IGFL2 | 147920 | 3 | 7 | 26 | |
| GSE74704 | IGFL2 | 147920 | 4 | 2 | 14 | |
| TCGA | IGFL2 | 147920 | 14 | 15 | 67 |
Total number of gains: 116; Total number of losses: 69; Total Number of normals: 303.
Somatic mutations of IGFL2:
Generating mutation plots.
Highly correlated genes for IGFL2:
Showing top 20/101 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| IGFL2 | CHGB | 0.811783 | 3 | 0 | 3 |
| IGFL2 | GALK2 | 0.784901 | 3 | 0 | 3 |
| IGFL2 | PPFIA4 | 0.772072 | 3 | 0 | 3 |
| IGFL2 | GLTPD2 | 0.750724 | 3 | 0 | 3 |
| IGFL2 | SCN2B | 0.734032 | 3 | 0 | 3 |
| IGFL2 | CRABP1 | 0.731794 | 3 | 0 | 3 |
| IGFL2 | SYCE3 | 0.728162 | 3 | 0 | 3 |
| IGFL2 | CDH2 | 0.723669 | 3 | 0 | 3 |
| IGFL2 | FZD9 | 0.708027 | 3 | 0 | 3 |
| IGFL2 | IGFN1 | 0.703308 | 3 | 0 | 3 |
| IGFL2 | FGG | 0.684371 | 3 | 0 | 3 |
| IGFL2 | TUBAL3 | 0.68077 | 4 | 0 | 3 |
| IGFL2 | FCN2 | 0.678876 | 4 | 0 | 4 |
| IGFL2 | CABP4 | 0.671954 | 3 | 0 | 3 |
| IGFL2 | THEG | 0.660384 | 3 | 0 | 3 |
| IGFL2 | SLC13A5 | 0.654549 | 4 | 0 | 3 |
| IGFL2 | TRPV3 | 0.64518 | 4 | 0 | 3 |
| IGFL2 | CHSY3 | 0.638545 | 5 | 0 | 4 |
| IGFL2 | PIK3CD | 0.632383 | 4 | 0 | 3 |
| IGFL2 | PDPN | 0.625178 | 3 | 0 | 3 |
For details and further investigation, click here