| Full name: histone acetyltransferase 1 | Alias Symbol: KAT1 | ||
| Type: protein-coding gene | Cytoband: 2q31.1 | ||
| Entrez ID: 8520 | HGNC ID: HGNC:4821 | Ensembl Gene: ENSG00000128708 | OMIM ID: 603053 |
| Drug and gene relationship at DGIdb | |||
Expression of HAT1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HAT1 | 8520 | 203138_at | 0.8210 | 0.0473 | |
| GSE20347 | HAT1 | 8520 | 203138_at | 1.0558 | 0.0001 | |
| GSE23400 | HAT1 | 8520 | 203138_at | 1.3588 | 0.0000 | |
| GSE26886 | HAT1 | 8520 | 203138_at | 0.5209 | 0.0308 | |
| GSE29001 | HAT1 | 8520 | 203138_at | 1.2928 | 0.0278 | |
| GSE38129 | HAT1 | 8520 | 203138_at | 1.1613 | 0.0000 | |
| GSE45670 | HAT1 | 8520 | 203138_at | 0.5622 | 0.0015 | |
| GSE53622 | HAT1 | 8520 | 69925 | 0.5049 | 0.0000 | |
| GSE53624 | HAT1 | 8520 | 69925 | 0.7000 | 0.0000 | |
| GSE63941 | HAT1 | 8520 | 203138_at | 0.9770 | 0.0043 | |
| GSE77861 | HAT1 | 8520 | 203138_at | 0.8064 | 0.0189 | |
| GSE97050 | HAT1 | 8520 | A_23_P339480 | 0.1652 | 0.6701 | |
| SRP007169 | HAT1 | 8520 | RNAseq | 1.1065 | 0.0030 | |
| SRP008496 | HAT1 | 8520 | RNAseq | 1.2103 | 0.0000 | |
| SRP064894 | HAT1 | 8520 | RNAseq | 0.5979 | 0.0007 | |
| SRP133303 | HAT1 | 8520 | RNAseq | 0.8935 | 0.0000 | |
| SRP159526 | HAT1 | 8520 | RNAseq | 0.4203 | 0.0697 | |
| SRP193095 | HAT1 | 8520 | RNAseq | 0.3085 | 0.0395 | |
| SRP219564 | HAT1 | 8520 | RNAseq | 0.3652 | 0.0679 | |
| TCGA | HAT1 | 8520 | RNAseq | 0.2067 | 0.0001 |
Upregulated datasets: 6; Downregulated datasets: 0.
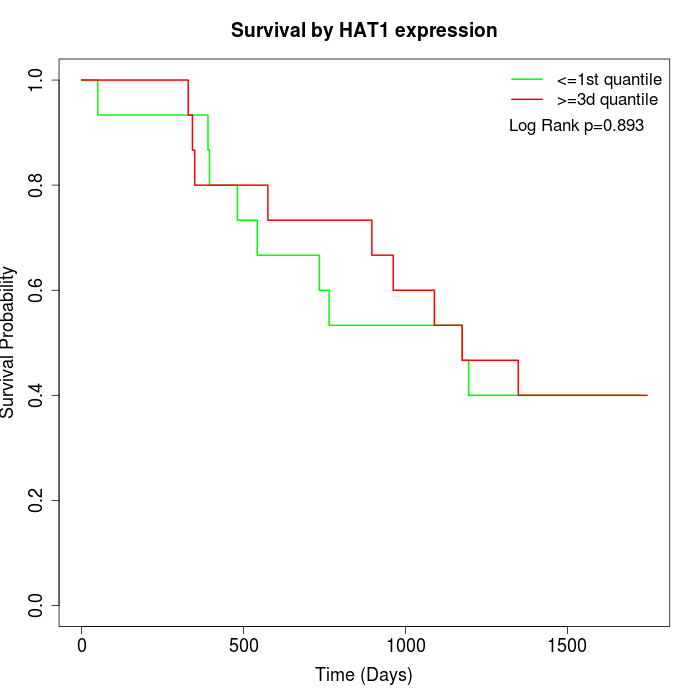
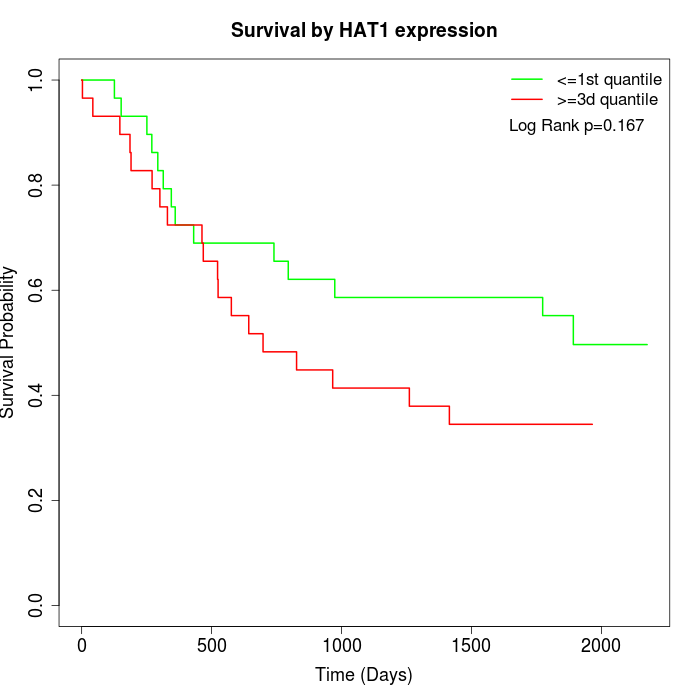
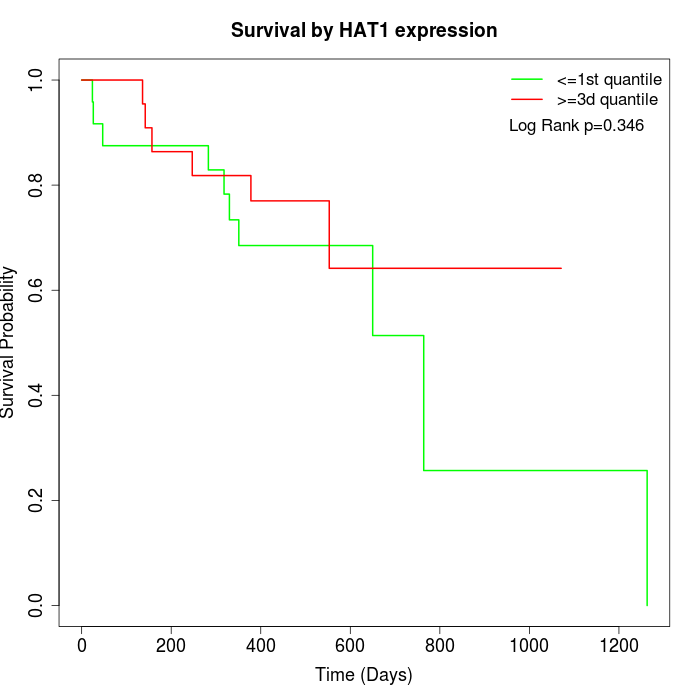
Survival by HAT1 expression:
Note: Click image to view full size file.
Copy number change of HAT1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HAT1 | 8520 | 8 | 0 | 22 | |
| GSE20123 | HAT1 | 8520 | 8 | 0 | 22 | |
| GSE43470 | HAT1 | 8520 | 6 | 1 | 36 | |
| GSE46452 | HAT1 | 8520 | 1 | 4 | 54 | |
| GSE47630 | HAT1 | 8520 | 5 | 3 | 32 | |
| GSE54993 | HAT1 | 8520 | 0 | 5 | 65 | |
| GSE54994 | HAT1 | 8520 | 11 | 3 | 39 | |
| GSE60625 | HAT1 | 8520 | 0 | 3 | 8 | |
| GSE74703 | HAT1 | 8520 | 5 | 1 | 30 | |
| GSE74704 | HAT1 | 8520 | 5 | 0 | 15 | |
| TCGA | HAT1 | 8520 | 28 | 6 | 62 |
Total number of gains: 77; Total number of losses: 26; Total Number of normals: 385.
Somatic mutations of HAT1:
Generating mutation plots.
Highly correlated genes for HAT1:
Showing top 20/1918 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HAT1 | CCDC15 | 0.768788 | 3 | 0 | 3 |
| HAT1 | TMEM185B | 0.754856 | 12 | 0 | 12 |
| HAT1 | ANAPC1 | 0.752075 | 3 | 0 | 3 |
| HAT1 | SPC25 | 0.748937 | 12 | 0 | 12 |
| HAT1 | AMMECR1L | 0.748215 | 6 | 0 | 6 |
| HAT1 | SSB | 0.746268 | 12 | 0 | 12 |
| HAT1 | PSMD14 | 0.743652 | 12 | 0 | 12 |
| HAT1 | PFDN2 | 0.742169 | 11 | 0 | 11 |
| HAT1 | MCM6 | 0.739457 | 13 | 0 | 13 |
| HAT1 | CENPBD1 | 0.737675 | 3 | 0 | 3 |
| HAT1 | SLC25A32 | 0.737398 | 11 | 0 | 11 |
| HAT1 | GMNN | 0.736426 | 13 | 0 | 12 |
| HAT1 | ATP6V1C1 | 0.735369 | 12 | 0 | 12 |
| HAT1 | ILF2 | 0.733922 | 12 | 0 | 12 |
| HAT1 | DNAJC10 | 0.733382 | 9 | 0 | 9 |
| HAT1 | CKS1B | 0.728915 | 13 | 0 | 12 |
| HAT1 | ARL6IP6 | 0.726075 | 9 | 0 | 9 |
| HAT1 | RRP15 | 0.723732 | 12 | 0 | 11 |
| HAT1 | PDCD2L | 0.722156 | 4 | 0 | 4 |
| HAT1 | UBE2E3 | 0.721797 | 12 | 0 | 11 |
For details and further investigation, click here