| Full name: hematopoietic cell signal transducer | Alias Symbol: DAP10|DKFZP586C1522|KAP10 | ||
| Type: protein-coding gene | Cytoband: 19q13.12 | ||
| Entrez ID: 10870 | HGNC ID: HGNC:16977 | Ensembl Gene: ENSG00000126264 | OMIM ID: 604089 |
| Drug and gene relationship at DGIdb | |||
HCST involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04650 | Natural killer cell mediated cytotoxicity |
Expression of HCST:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HCST | 10870 | 223640_at | 0.0309 | 0.9744 | |
| GSE26886 | HCST | 10870 | 223640_at | -0.1538 | 0.5434 | |
| GSE45670 | HCST | 10870 | 223640_at | -0.2286 | 0.1323 | |
| GSE53622 | HCST | 10870 | 65686 | -0.1383 | 0.3555 | |
| GSE53624 | HCST | 10870 | 65686 | -0.4205 | 0.0002 | |
| GSE63941 | HCST | 10870 | 223640_at | -0.0033 | 0.9885 | |
| GSE77861 | HCST | 10870 | 223640_at | -0.2434 | 0.1015 | |
| GSE97050 | HCST | 10870 | A_23_P39386 | 0.6097 | 0.2368 | |
| SRP064894 | HCST | 10870 | RNAseq | 1.4982 | 0.0053 | |
| SRP133303 | HCST | 10870 | RNAseq | -0.0635 | 0.8421 | |
| SRP159526 | HCST | 10870 | RNAseq | -0.4764 | 0.5261 | |
| SRP193095 | HCST | 10870 | RNAseq | -0.2552 | 0.2166 | |
| SRP219564 | HCST | 10870 | RNAseq | 0.9219 | 0.1322 | |
| TCGA | HCST | 10870 | RNAseq | 0.1722 | 0.4071 |
Upregulated datasets: 1; Downregulated datasets: 0.
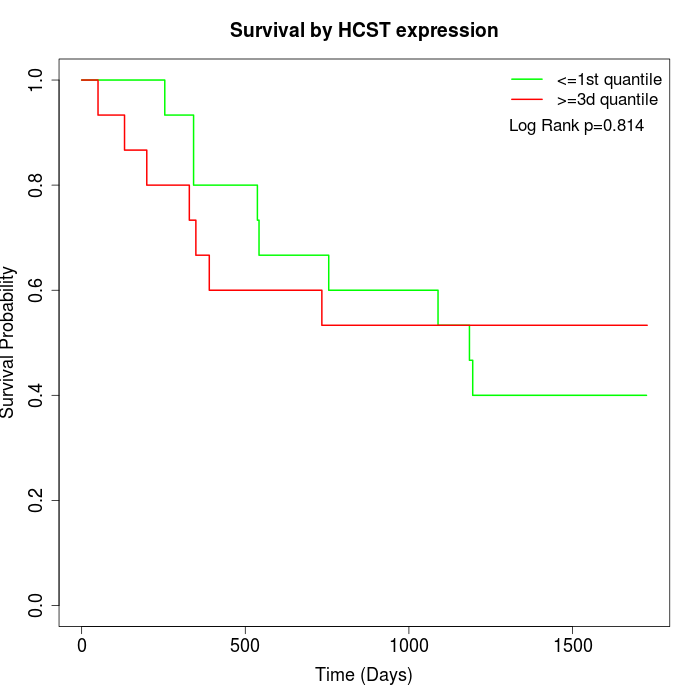
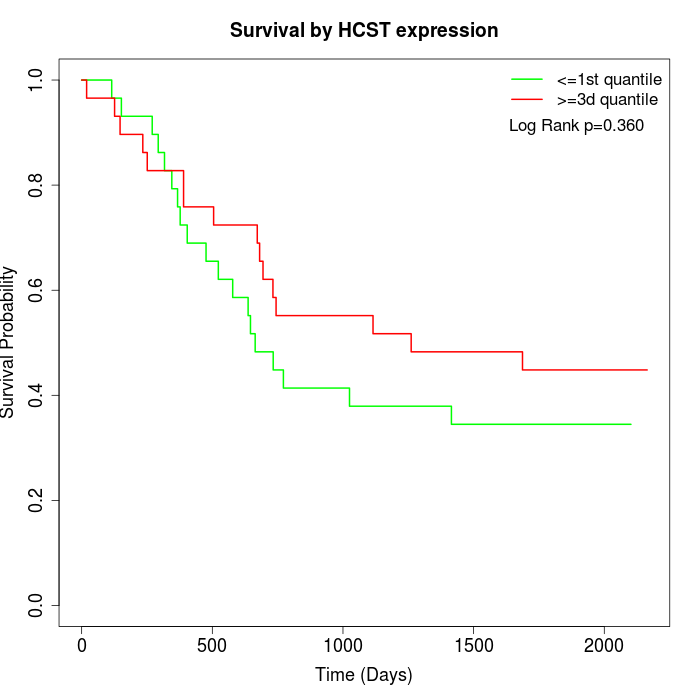
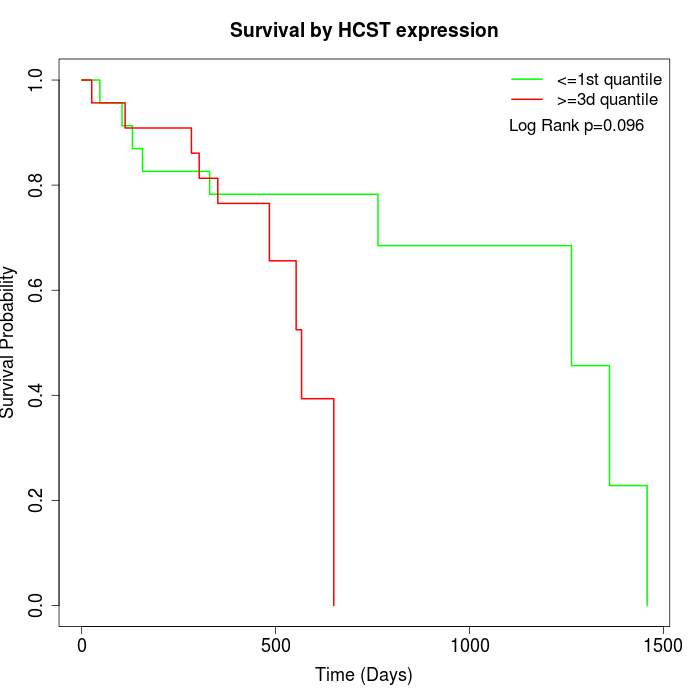
Survival by HCST expression:
Note: Click image to view full size file.
Copy number change of HCST:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HCST | 10870 | 7 | 4 | 19 | |
| GSE20123 | HCST | 10870 | 7 | 3 | 20 | |
| GSE43470 | HCST | 10870 | 3 | 7 | 33 | |
| GSE46452 | HCST | 10870 | 48 | 1 | 10 | |
| GSE47630 | HCST | 10870 | 9 | 5 | 26 | |
| GSE54993 | HCST | 10870 | 17 | 3 | 50 | |
| GSE54994 | HCST | 10870 | 6 | 9 | 38 | |
| GSE60625 | HCST | 10870 | 9 | 0 | 2 | |
| GSE74703 | HCST | 10870 | 3 | 4 | 29 | |
| GSE74704 | HCST | 10870 | 6 | 1 | 13 | |
| TCGA | HCST | 10870 | 22 | 10 | 64 |
Total number of gains: 137; Total number of losses: 47; Total Number of normals: 304.
Somatic mutations of HCST:
Generating mutation plots.
Highly correlated genes for HCST:
Showing top 20/324 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HCST | AIF1 | 0.783851 | 7 | 0 | 7 |
| HCST | CYBB | 0.768437 | 6 | 0 | 6 |
| HCST | CD53 | 0.741266 | 5 | 0 | 5 |
| HCST | CYTIP | 0.730886 | 5 | 0 | 5 |
| HCST | CD3D | 0.729995 | 8 | 0 | 8 |
| HCST | RNASE6 | 0.722694 | 6 | 0 | 6 |
| HCST | TNFSF8 | 0.721312 | 4 | 0 | 4 |
| HCST | CD52 | 0.712517 | 8 | 0 | 7 |
| HCST | C1orf162 | 0.704162 | 7 | 0 | 7 |
| HCST | NCKAP1L | 0.702379 | 6 | 0 | 5 |
| HCST | CXCR6 | 0.701313 | 4 | 0 | 4 |
| HCST | SLA | 0.700475 | 7 | 0 | 7 |
| HCST | CD74 | 0.699002 | 8 | 0 | 8 |
| HCST | CD37 | 0.698994 | 5 | 0 | 4 |
| HCST | ITGB2 | 0.698909 | 7 | 0 | 6 |
| HCST | GMFG | 0.697955 | 7 | 0 | 7 |
| HCST | LAPTM5 | 0.696283 | 7 | 0 | 7 |
| HCST | GIMAP2 | 0.693833 | 7 | 0 | 6 |
| HCST | HLA-DMB | 0.691448 | 7 | 0 | 7 |
| HCST | SASH3 | 0.691195 | 7 | 0 | 6 |
For details and further investigation, click here