| Full name: integrin subunit beta 2 | Alias Symbol: LFA-1|MAC-1 | ||
| Type: protein-coding gene | Cytoband: 21q22.3 | ||
| Entrez ID: 3689 | HGNC ID: HGNC:6155 | Ensembl Gene: ENSG00000160255 | OMIM ID: 600065 |
| Drug and gene relationship at DGIdb | |||
ITGB2 involved pathways:
Expression of ITGB2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ITGB2 | 3689 | 202803_s_at | 0.7508 | 0.5909 | |
| GSE20347 | ITGB2 | 3689 | 202803_s_at | 0.1267 | 0.6146 | |
| GSE23400 | ITGB2 | 3689 | 202803_s_at | 0.4511 | 0.0001 | |
| GSE26886 | ITGB2 | 3689 | 202803_s_at | -0.1589 | 0.7528 | |
| GSE29001 | ITGB2 | 3689 | 202803_s_at | 0.3740 | 0.1445 | |
| GSE38129 | ITGB2 | 3689 | 202803_s_at | 0.3139 | 0.2752 | |
| GSE45670 | ITGB2 | 3689 | 202803_s_at | 0.1075 | 0.7620 | |
| GSE53622 | ITGB2 | 3689 | 15347 | 0.5315 | 0.0003 | |
| GSE53624 | ITGB2 | 3689 | 96687 | 0.5693 | 0.0000 | |
| GSE63941 | ITGB2 | 3689 | 236988_x_at | 0.1294 | 0.3810 | |
| GSE77861 | ITGB2 | 3689 | 236988_x_at | 0.0080 | 0.9736 | |
| GSE97050 | ITGB2 | 3689 | A_23_P430411 | 1.1869 | 0.0782 | |
| SRP007169 | ITGB2 | 3689 | RNAseq | 2.9028 | 0.0012 | |
| SRP008496 | ITGB2 | 3689 | RNAseq | 0.3135 | 0.3989 | |
| SRP064894 | ITGB2 | 3689 | RNAseq | 1.6083 | 0.0000 | |
| SRP133303 | ITGB2 | 3689 | RNAseq | 0.6488 | 0.0079 | |
| SRP159526 | ITGB2 | 3689 | RNAseq | 0.2953 | 0.6887 | |
| SRP193095 | ITGB2 | 3689 | RNAseq | 0.6047 | 0.0035 | |
| SRP219564 | ITGB2 | 3689 | RNAseq | 1.4224 | 0.0199 | |
| TCGA | ITGB2 | 3689 | RNAseq | 0.3768 | 0.0014 |
Upregulated datasets: 3; Downregulated datasets: 0.
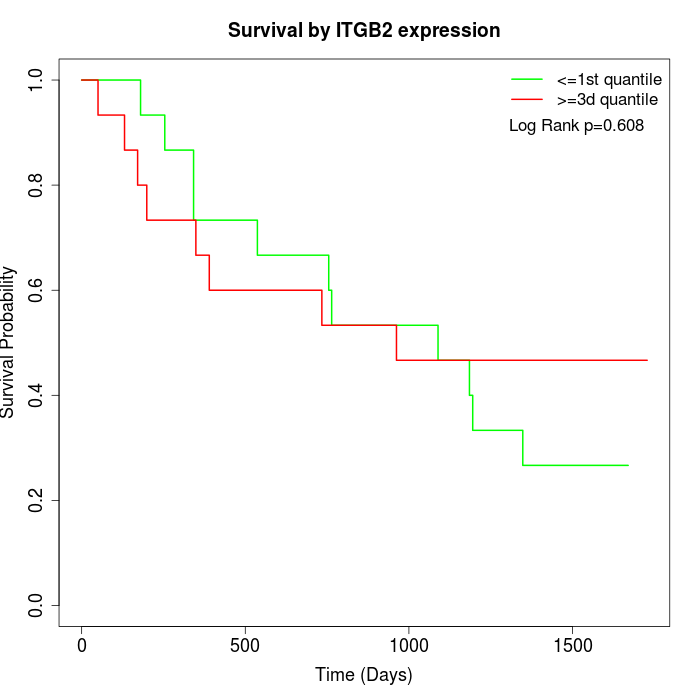
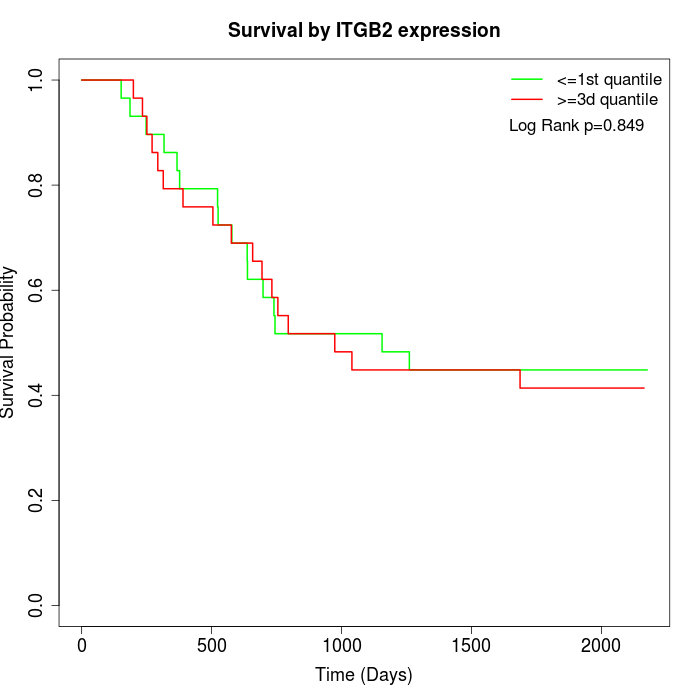
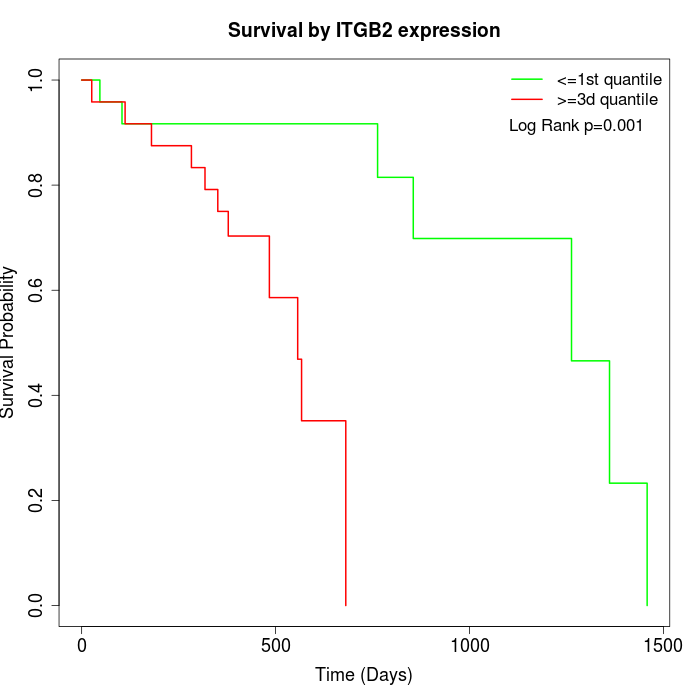
Survival by ITGB2 expression:
Note: Click image to view full size file.
Copy number change of ITGB2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ITGB2 | 3689 | 2 | 9 | 19 | |
| GSE20123 | ITGB2 | 3689 | 2 | 9 | 19 | |
| GSE43470 | ITGB2 | 3689 | 0 | 13 | 30 | |
| GSE46452 | ITGB2 | 3689 | 1 | 21 | 37 | |
| GSE47630 | ITGB2 | 3689 | 6 | 17 | 17 | |
| GSE54993 | ITGB2 | 3689 | 8 | 1 | 61 | |
| GSE54994 | ITGB2 | 3689 | 4 | 8 | 41 | |
| GSE60625 | ITGB2 | 3689 | 0 | 0 | 11 | |
| GSE74703 | ITGB2 | 3689 | 0 | 10 | 26 | |
| GSE74704 | ITGB2 | 3689 | 1 | 5 | 14 | |
| TCGA | ITGB2 | 3689 | 9 | 38 | 49 |
Total number of gains: 33; Total number of losses: 131; Total Number of normals: 324.
Somatic mutations of ITGB2:
Generating mutation plots.
Highly correlated genes for ITGB2:
Showing top 20/557 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ITGB2 | CD53 | 0.831958 | 9 | 0 | 9 |
| ITGB2 | TYROBP | 0.815179 | 11 | 0 | 11 |
| ITGB2 | FCER1G | 0.805621 | 10 | 0 | 10 |
| ITGB2 | LAPTM5 | 0.803703 | 12 | 0 | 11 |
| ITGB2 | C1QC | 0.7821 | 7 | 0 | 7 |
| ITGB2 | AIF1 | 0.774749 | 11 | 0 | 11 |
| ITGB2 | GRAPL | 0.758586 | 3 | 0 | 3 |
| ITGB2 | DERL3 | 0.756583 | 3 | 0 | 3 |
| ITGB2 | CYBB | 0.754686 | 11 | 0 | 11 |
| ITGB2 | HAVCR2 | 0.75233 | 6 | 0 | 5 |
| ITGB2 | C1QA | 0.752128 | 11 | 0 | 11 |
| ITGB2 | TFEC | 0.749196 | 11 | 0 | 10 |
| ITGB2 | NCKAP1L | 0.749092 | 10 | 0 | 9 |
| ITGB2 | FCGR3A | 0.747457 | 3 | 0 | 3 |
| ITGB2 | ITGB2-AS1 | 0.744815 | 4 | 0 | 4 |
| ITGB2 | C1QB | 0.743569 | 11 | 0 | 11 |
| ITGB2 | PLEK | 0.743132 | 5 | 0 | 5 |
| ITGB2 | LCP2 | 0.73528 | 10 | 0 | 9 |
| ITGB2 | RASSF4 | 0.729635 | 6 | 0 | 6 |
| ITGB2 | TNFRSF1B | 0.729275 | 12 | 0 | 11 |
For details and further investigation, click here