| Full name: histone deacetylase 2 | Alias Symbol: RPD3|YAF1|KDAC2 | ||
| Type: protein-coding gene | Cytoband: 6q21 | ||
| Entrez ID: 3066 | HGNC ID: HGNC:4853 | Ensembl Gene: ENSG00000196591 | OMIM ID: 605164 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
HDAC2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04110 | Cell cycle | |
| hsa04213 | Longevity regulating pathway - multiple species | |
| hsa04330 | Notch signaling pathway | |
| hsa05200 | Pathways in cancer | |
| hsa05220 | Chronic myeloid leukemia |
Expression of HDAC2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HDAC2 | 3066 | 201833_at | 0.1966 | 0.6496 | |
| GSE20347 | HDAC2 | 3066 | 201833_at | 0.4149 | 0.0729 | |
| GSE23400 | HDAC2 | 3066 | 201833_at | 0.5169 | 0.0000 | |
| GSE26886 | HDAC2 | 3066 | 201833_at | 0.4020 | 0.0769 | |
| GSE29001 | HDAC2 | 3066 | 201833_at | 0.3531 | 0.4336 | |
| GSE38129 | HDAC2 | 3066 | 201833_at | 0.5171 | 0.0003 | |
| GSE45670 | HDAC2 | 3066 | 201833_at | 0.4115 | 0.0010 | |
| GSE53622 | HDAC2 | 3066 | 32592 | 0.4343 | 0.0000 | |
| GSE53624 | HDAC2 | 3066 | 32592 | 0.5363 | 0.0000 | |
| GSE63941 | HDAC2 | 3066 | 201833_at | -0.4301 | 0.1802 | |
| GSE77861 | HDAC2 | 3066 | 201833_at | 0.4800 | 0.1166 | |
| GSE97050 | HDAC2 | 3066 | A_23_P122304 | 0.3563 | 0.2501 | |
| SRP007169 | HDAC2 | 3066 | RNAseq | 1.7013 | 0.0000 | |
| SRP008496 | HDAC2 | 3066 | RNAseq | 1.7518 | 0.0000 | |
| SRP064894 | HDAC2 | 3066 | RNAseq | 0.3361 | 0.0242 | |
| SRP133303 | HDAC2 | 3066 | RNAseq | 0.5112 | 0.0003 | |
| SRP159526 | HDAC2 | 3066 | RNAseq | 0.4964 | 0.0051 | |
| SRP193095 | HDAC2 | 3066 | RNAseq | 0.0140 | 0.8857 | |
| SRP219564 | HDAC2 | 3066 | RNAseq | 0.0035 | 0.9885 | |
| TCGA | HDAC2 | 3066 | RNAseq | 0.1203 | 0.0085 |
Upregulated datasets: 2; Downregulated datasets: 0.
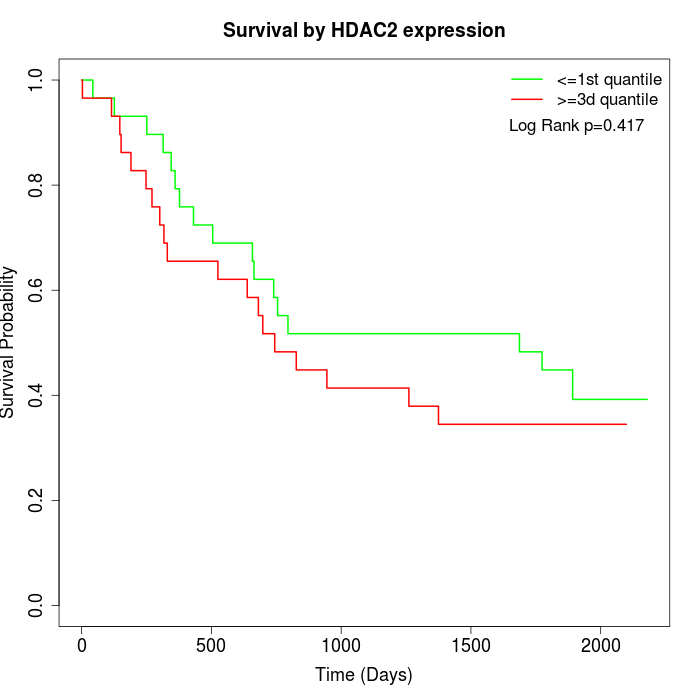
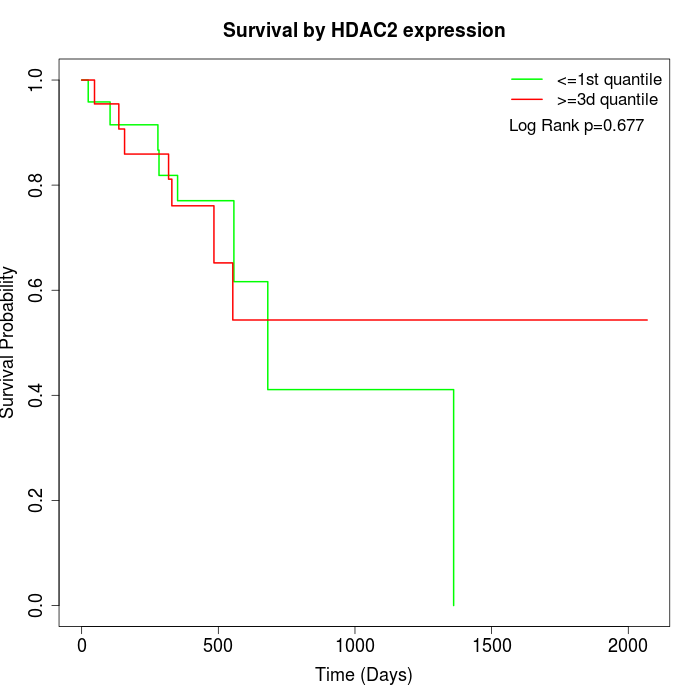
Survival by HDAC2 expression:
Note: Click image to view full size file.
Copy number change of HDAC2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HDAC2 | 3066 | 1 | 4 | 25 | |
| GSE20123 | HDAC2 | 3066 | 1 | 3 | 26 | |
| GSE43470 | HDAC2 | 3066 | 3 | 1 | 39 | |
| GSE46452 | HDAC2 | 3066 | 2 | 11 | 46 | |
| GSE47630 | HDAC2 | 3066 | 9 | 4 | 27 | |
| GSE54993 | HDAC2 | 3066 | 3 | 2 | 65 | |
| GSE54994 | HDAC2 | 3066 | 8 | 8 | 37 | |
| GSE60625 | HDAC2 | 3066 | 0 | 2 | 9 | |
| GSE74703 | HDAC2 | 3066 | 2 | 1 | 33 | |
| GSE74704 | HDAC2 | 3066 | 0 | 1 | 19 | |
| TCGA | HDAC2 | 3066 | 7 | 22 | 67 |
Total number of gains: 36; Total number of losses: 59; Total Number of normals: 393.
Somatic mutations of HDAC2:
Generating mutation plots.
Highly correlated genes for HDAC2:
Showing top 20/864 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HDAC2 | MMS22L | 0.742872 | 3 | 0 | 3 |
| HDAC2 | PSME1 | 0.732656 | 3 | 0 | 3 |
| HDAC2 | WIPF1 | 0.729745 | 3 | 0 | 3 |
| HDAC2 | ANAPC1 | 0.7218 | 3 | 0 | 3 |
| HDAC2 | PYCR2 | 0.716529 | 3 | 0 | 3 |
| HDAC2 | RPF2 | 0.714474 | 7 | 0 | 6 |
| HDAC2 | POLR3F | 0.70351 | 8 | 0 | 8 |
| HDAC2 | PPIG | 0.703443 | 6 | 0 | 6 |
| HDAC2 | TCP1 | 0.698385 | 4 | 0 | 4 |
| HDAC2 | NEB | 0.693867 | 3 | 0 | 3 |
| HDAC2 | PPP1R2 | 0.691928 | 3 | 0 | 3 |
| HDAC2 | FASTKD1 | 0.690726 | 4 | 0 | 3 |
| HDAC2 | PTCD1 | 0.682174 | 3 | 0 | 3 |
| HDAC2 | FIP1L1 | 0.677629 | 6 | 0 | 4 |
| HDAC2 | NBEAL1 | 0.677387 | 3 | 0 | 3 |
| HDAC2 | SUMO2 | 0.67548 | 3 | 0 | 3 |
| HDAC2 | EIF3L | 0.674544 | 3 | 0 | 3 |
| HDAC2 | SSB | 0.674106 | 12 | 0 | 11 |
| HDAC2 | SPIC | 0.671675 | 3 | 0 | 3 |
| HDAC2 | TSN | 0.669838 | 7 | 0 | 7 |
For details and further investigation, click here