| Full name: heparan sulfate-glucosamine 3-sulfotransferase 4 | Alias Symbol: 3OST4 | ||
| Type: protein-coding gene | Cytoband: 16p12.1 | ||
| Entrez ID: 9951 | HGNC ID: HGNC:5200 | Ensembl Gene: ENSG00000182601 | OMIM ID: 604059 |
| Drug and gene relationship at DGIdb | |||
Expression of HS3ST4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HS3ST4 | 9951 | 228206_at | -0.4413 | 0.2966 | |
| GSE26886 | HS3ST4 | 9951 | 228206_at | -0.1763 | 0.0533 | |
| GSE45670 | HS3ST4 | 9951 | 228206_at | 0.1334 | 0.7335 | |
| GSE53622 | HS3ST4 | 9951 | 60542 | -1.3247 | 0.0000 | |
| GSE53624 | HS3ST4 | 9951 | 60542 | -2.6695 | 0.0000 | |
| GSE63941 | HS3ST4 | 9951 | 228206_at | 0.2504 | 0.5489 | |
| GSE77861 | HS3ST4 | 9951 | 228206_at | -0.1090 | 0.3073 | |
| SRP133303 | HS3ST4 | 9951 | RNAseq | -2.2679 | 0.0001 | |
| SRP159526 | HS3ST4 | 9951 | RNAseq | -0.6909 | 0.4001 | |
| TCGA | HS3ST4 | 9951 | RNAseq | -1.8342 | 0.0311 |
Upregulated datasets: 0; Downregulated datasets: 4.
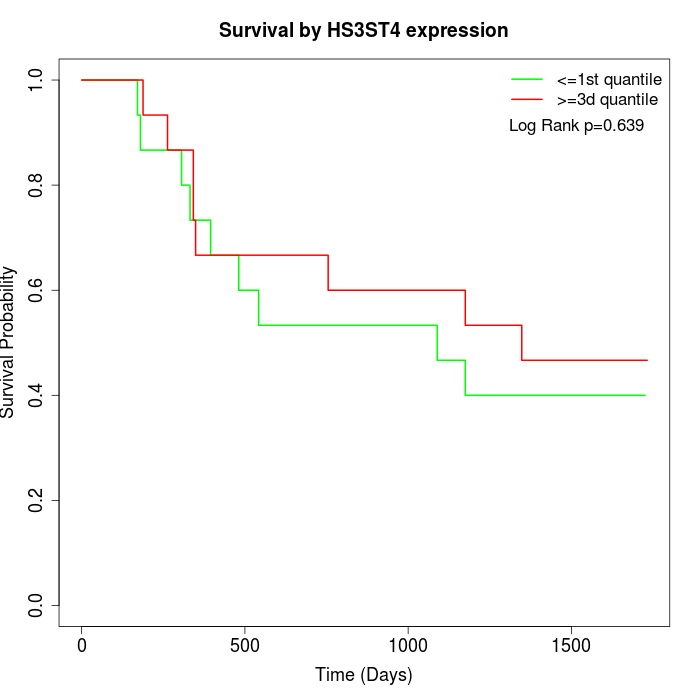
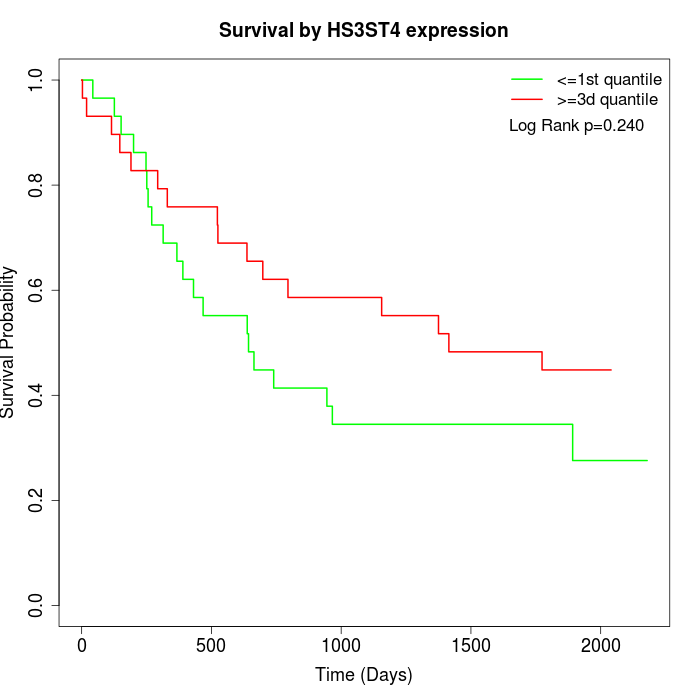
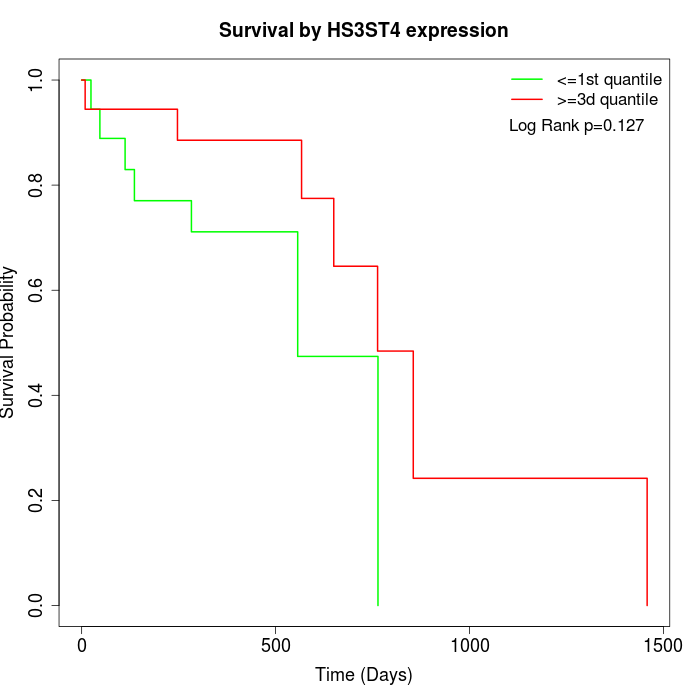
Survival by HS3ST4 expression:
Note: Click image to view full size file.
Copy number change of HS3ST4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HS3ST4 | 9951 | 4 | 3 | 23 | |
| GSE20123 | HS3ST4 | 9951 | 4 | 2 | 24 | |
| GSE43470 | HS3ST4 | 9951 | 4 | 2 | 37 | |
| GSE46452 | HS3ST4 | 9951 | 38 | 1 | 20 | |
| GSE47630 | HS3ST4 | 9951 | 12 | 6 | 22 | |
| GSE54993 | HS3ST4 | 9951 | 3 | 5 | 62 | |
| GSE54994 | HS3ST4 | 9951 | 4 | 9 | 40 | |
| GSE60625 | HS3ST4 | 9951 | 4 | 0 | 7 | |
| GSE74703 | HS3ST4 | 9951 | 4 | 2 | 30 | |
| GSE74704 | HS3ST4 | 9951 | 2 | 1 | 17 | |
| TCGA | HS3ST4 | 9951 | 19 | 13 | 64 |
Total number of gains: 98; Total number of losses: 44; Total Number of normals: 346.
Somatic mutations of HS3ST4:
Generating mutation plots.
Highly correlated genes for HS3ST4:
Showing top 20/56 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HS3ST4 | EFNA2 | 0.691706 | 3 | 0 | 3 |
| HS3ST4 | BEAN1 | 0.639646 | 3 | 0 | 3 |
| HS3ST4 | SAMD10 | 0.607954 | 5 | 0 | 4 |
| HS3ST4 | AVPR1B | 0.607383 | 3 | 0 | 3 |
| HS3ST4 | SPATA31E1 | 0.603027 | 3 | 0 | 3 |
| HS3ST4 | SLC25A51 | 0.583586 | 3 | 0 | 3 |
| HS3ST4 | LRRC3B | 0.574692 | 4 | 0 | 3 |
| HS3ST4 | MTA2 | 0.573833 | 3 | 0 | 3 |
| HS3ST4 | ARSD | 0.571248 | 4 | 0 | 3 |
| HS3ST4 | LMO7 | 0.566432 | 4 | 0 | 3 |
| HS3ST4 | FAM189A2 | 0.562899 | 3 | 0 | 3 |
| HS3ST4 | CXCR2 | 0.556567 | 6 | 0 | 3 |
| HS3ST4 | CACNB4 | 0.553769 | 5 | 0 | 4 |
| HS3ST4 | CLEC4F | 0.550004 | 4 | 0 | 3 |
| HS3ST4 | TMEM114 | 0.547768 | 3 | 0 | 3 |
| HS3ST4 | GCKR | 0.542947 | 5 | 0 | 3 |
| HS3ST4 | TP53I3 | 0.542828 | 5 | 0 | 3 |
| HS3ST4 | NCOA1 | 0.541925 | 4 | 0 | 3 |
| HS3ST4 | KALRN | 0.541458 | 5 | 0 | 3 |
| HS3ST4 | AHSP | 0.540268 | 4 | 0 | 3 |
For details and further investigation, click here