| Full name: heparan sulfate proteoglycan 2 | Alias Symbol: perlecan|PRCAN | ||
| Type: protein-coding gene | Cytoband: 1p36.12 | ||
| Entrez ID: 3339 | HGNC ID: HGNC:5273 | Ensembl Gene: ENSG00000142798 | OMIM ID: 142461 |
| Drug and gene relationship at DGIdb | |||
HSPG2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05205 | Proteoglycans in cancer |
Expression of HSPG2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HSPG2 | 3339 | 201655_s_at | 0.5183 | 0.4826 | |
| GSE20347 | HSPG2 | 3339 | 201655_s_at | 0.5550 | 0.0082 | |
| GSE23400 | HSPG2 | 3339 | 201655_s_at | 0.1905 | 0.0962 | |
| GSE26886 | HSPG2 | 3339 | 201655_s_at | 1.4330 | 0.0000 | |
| GSE29001 | HSPG2 | 3339 | 201655_s_at | 0.4240 | 0.0290 | |
| GSE38129 | HSPG2 | 3339 | 201655_s_at | 0.1072 | 0.7576 | |
| GSE45670 | HSPG2 | 3339 | 201655_s_at | -0.1672 | 0.5065 | |
| GSE53622 | HSPG2 | 3339 | 63266 | 0.3371 | 0.0003 | |
| GSE53624 | HSPG2 | 3339 | 63266 | 0.5791 | 0.0000 | |
| GSE63941 | HSPG2 | 3339 | 201655_s_at | -1.5962 | 0.0260 | |
| GSE77861 | HSPG2 | 3339 | 201655_s_at | 0.6254 | 0.0465 | |
| GSE97050 | HSPG2 | 3339 | A_33_P3380612 | 0.0636 | 0.8409 | |
| SRP007169 | HSPG2 | 3339 | RNAseq | 2.6873 | 0.0000 | |
| SRP008496 | HSPG2 | 3339 | RNAseq | 2.6998 | 0.0000 | |
| SRP064894 | HSPG2 | 3339 | RNAseq | 0.4829 | 0.1656 | |
| SRP133303 | HSPG2 | 3339 | RNAseq | 0.3130 | 0.3019 | |
| SRP159526 | HSPG2 | 3339 | RNAseq | -0.2268 | 0.4080 | |
| SRP193095 | HSPG2 | 3339 | RNAseq | 1.3696 | 0.0000 | |
| SRP219564 | HSPG2 | 3339 | RNAseq | -0.3341 | 0.5816 | |
| TCGA | HSPG2 | 3339 | RNAseq | -0.2092 | 0.0018 |
Upregulated datasets: 4; Downregulated datasets: 1.
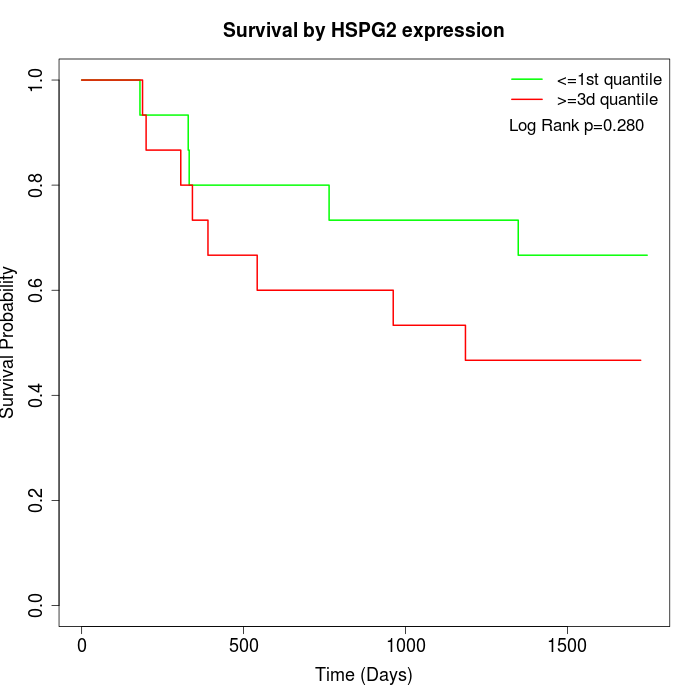
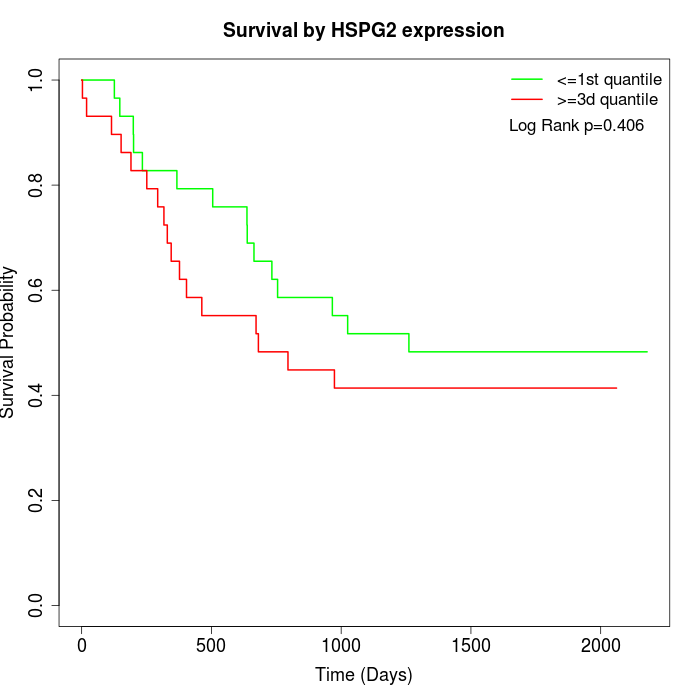
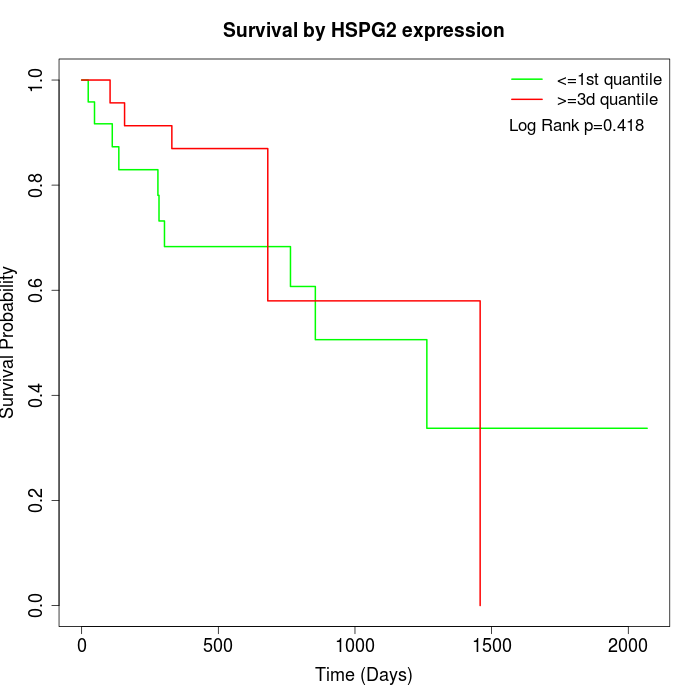
Survival by HSPG2 expression:
Note: Click image to view full size file.
Copy number change of HSPG2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HSPG2 | 3339 | 0 | 5 | 25 | |
| GSE20123 | HSPG2 | 3339 | 0 | 4 | 26 | |
| GSE43470 | HSPG2 | 3339 | 2 | 6 | 35 | |
| GSE46452 | HSPG2 | 3339 | 5 | 1 | 53 | |
| GSE47630 | HSPG2 | 3339 | 8 | 3 | 29 | |
| GSE54993 | HSPG2 | 3339 | 2 | 1 | 67 | |
| GSE54994 | HSPG2 | 3339 | 11 | 4 | 38 | |
| GSE60625 | HSPG2 | 3339 | 0 | 0 | 11 | |
| GSE74703 | HSPG2 | 3339 | 1 | 4 | 31 | |
| GSE74704 | HSPG2 | 3339 | 0 | 0 | 20 | |
| TCGA | HSPG2 | 3339 | 9 | 23 | 64 |
Total number of gains: 38; Total number of losses: 51; Total Number of normals: 399.
Somatic mutations of HSPG2:
Generating mutation plots.
Highly correlated genes for HSPG2:
Showing top 20/1434 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HSPG2 | SLC39A3 | 0.837168 | 3 | 0 | 3 |
| HSPG2 | TP53I13 | 0.771432 | 5 | 0 | 5 |
| HSPG2 | SYNC | 0.765055 | 4 | 0 | 4 |
| HSPG2 | SLC39A13 | 0.763391 | 8 | 0 | 8 |
| HSPG2 | CRTC2 | 0.747151 | 5 | 0 | 5 |
| HSPG2 | ZFYVE9 | 0.726847 | 3 | 0 | 3 |
| HSPG2 | MFGE8 | 0.725019 | 12 | 0 | 12 |
| HSPG2 | KRTCAP2 | 0.722008 | 4 | 0 | 4 |
| HSPG2 | MNT | 0.721754 | 3 | 0 | 3 |
| HSPG2 | NACC2 | 0.719885 | 4 | 0 | 4 |
| HSPG2 | TRIM47 | 0.718828 | 5 | 0 | 4 |
| HSPG2 | SCYL1 | 0.71778 | 3 | 0 | 3 |
| HSPG2 | ARMC9 | 0.717357 | 7 | 0 | 6 |
| HSPG2 | SSC5D | 0.716967 | 3 | 0 | 3 |
| HSPG2 | FUBP1 | 0.716766 | 3 | 0 | 3 |
| HSPG2 | SPIRE1 | 0.716711 | 4 | 0 | 4 |
| HSPG2 | EHBP1L1 | 0.714893 | 3 | 0 | 3 |
| HSPG2 | SLC25A42 | 0.710727 | 3 | 0 | 3 |
| HSPG2 | AGTR1 | 0.708032 | 4 | 0 | 4 |
| HSPG2 | THEM4 | 0.70768 | 3 | 0 | 3 |
For details and further investigation, click here