| Full name: calcium homeostasis modulator family member 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 10q24.33 | ||
| Entrez ID: 51063 | HGNC ID: HGNC:23493 | Ensembl Gene: ENSG00000138172 | OMIM ID: 612235 |
| Drug and gene relationship at DGIdb | |||
Expression of CALHM2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CALHM2 | 51063 | 221565_s_at | -0.0970 | 0.9230 | |
| GSE20347 | CALHM2 | 51063 | 221565_s_at | 0.2474 | 0.1312 | |
| GSE23400 | CALHM2 | 51063 | 221565_s_at | -0.0664 | 0.2809 | |
| GSE26886 | CALHM2 | 51063 | 221565_s_at | 0.5770 | 0.0287 | |
| GSE29001 | CALHM2 | 51063 | 221565_s_at | 0.3992 | 0.1596 | |
| GSE38129 | CALHM2 | 51063 | 221565_s_at | -0.0612 | 0.7717 | |
| GSE45670 | CALHM2 | 51063 | 57715_at | -0.7301 | 0.0033 | |
| GSE53622 | CALHM2 | 51063 | 58677 | 0.0386 | 0.7527 | |
| GSE53624 | CALHM2 | 51063 | 58677 | 0.2280 | 0.0144 | |
| GSE63941 | CALHM2 | 51063 | 221565_s_at | -1.7509 | 0.0211 | |
| GSE77861 | CALHM2 | 51063 | 221565_s_at | 0.0552 | 0.7970 | |
| GSE97050 | CALHM2 | 51063 | A_23_P202219 | -0.0012 | 0.9975 | |
| SRP007169 | CALHM2 | 51063 | RNAseq | 1.1712 | 0.0483 | |
| SRP008496 | CALHM2 | 51063 | RNAseq | 1.3142 | 0.0047 | |
| SRP064894 | CALHM2 | 51063 | RNAseq | 0.3355 | 0.3386 | |
| SRP133303 | CALHM2 | 51063 | RNAseq | -0.2079 | 0.3007 | |
| SRP159526 | CALHM2 | 51063 | RNAseq | 0.2453 | 0.4796 | |
| SRP193095 | CALHM2 | 51063 | RNAseq | 0.8201 | 0.0000 | |
| SRP219564 | CALHM2 | 51063 | RNAseq | 0.1569 | 0.7428 | |
| TCGA | CALHM2 | 51063 | RNAseq | 0.0257 | 0.7955 |
Upregulated datasets: 2; Downregulated datasets: 1.
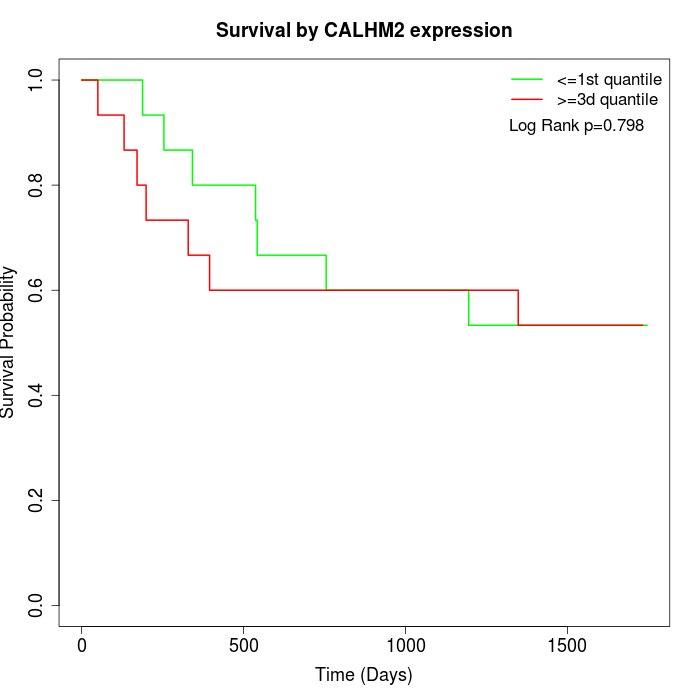
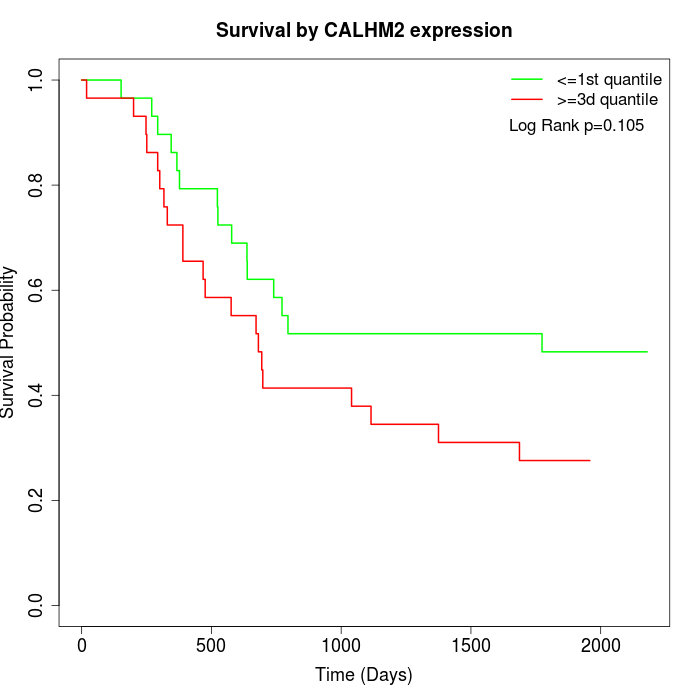
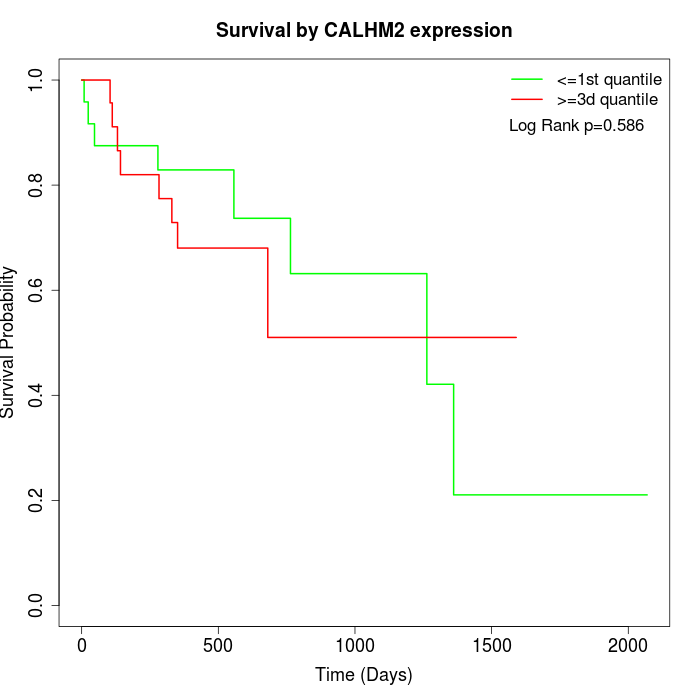
Survival by CALHM2 expression:
Note: Click image to view full size file.
Copy number change of CALHM2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CALHM2 | 51063 | 2 | 9 | 19 | |
| GSE20123 | CALHM2 | 51063 | 2 | 8 | 20 | |
| GSE43470 | CALHM2 | 51063 | 0 | 8 | 35 | |
| GSE46452 | CALHM2 | 51063 | 0 | 11 | 48 | |
| GSE47630 | CALHM2 | 51063 | 2 | 14 | 24 | |
| GSE54993 | CALHM2 | 51063 | 8 | 0 | 62 | |
| GSE54994 | CALHM2 | 51063 | 1 | 10 | 42 | |
| GSE60625 | CALHM2 | 51063 | 0 | 0 | 11 | |
| GSE74703 | CALHM2 | 51063 | 0 | 5 | 31 | |
| GSE74704 | CALHM2 | 51063 | 1 | 4 | 15 | |
| TCGA | CALHM2 | 51063 | 6 | 28 | 62 |
Total number of gains: 22; Total number of losses: 97; Total Number of normals: 369.
Somatic mutations of CALHM2:
Generating mutation plots.
Highly correlated genes for CALHM2:
Showing top 20/617 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CALHM2 | CLEC14A | 0.777873 | 3 | 0 | 3 |
| CALHM2 | ANKRD44 | 0.70227 | 3 | 0 | 3 |
| CALHM2 | PMP22 | 0.694096 | 9 | 0 | 9 |
| CALHM2 | ICAM2 | 0.690076 | 5 | 0 | 5 |
| CALHM2 | DCHS1 | 0.677324 | 9 | 0 | 8 |
| CALHM2 | BNC2 | 0.672307 | 7 | 0 | 7 |
| CALHM2 | NRROS | 0.671988 | 5 | 0 | 4 |
| CALHM2 | SDC2 | 0.669153 | 8 | 0 | 8 |
| CALHM2 | GPC6 | 0.668899 | 5 | 0 | 5 |
| CALHM2 | ITIH5 | 0.665221 | 5 | 0 | 4 |
| CALHM2 | EPHA3 | 0.661537 | 5 | 0 | 5 |
| CALHM2 | UNC5C | 0.660907 | 4 | 0 | 4 |
| CALHM2 | COLEC12 | 0.659395 | 9 | 0 | 9 |
| CALHM2 | DPYSL2 | 0.656947 | 10 | 0 | 9 |
| CALHM2 | CNRIP1 | 0.653241 | 6 | 0 | 5 |
| CALHM2 | BHLHE22 | 0.6501 | 4 | 0 | 4 |
| CALHM2 | FXYD6 | 0.649509 | 10 | 0 | 8 |
| CALHM2 | SLC16A4 | 0.648592 | 8 | 0 | 8 |
| CALHM2 | DDR2 | 0.646345 | 10 | 0 | 9 |
| CALHM2 | ITGAM | 0.644666 | 7 | 0 | 6 |
For details and further investigation, click here