| Full name: interleukin 1 receptor type 2 | Alias Symbol: CD121b | ||
| Type: protein-coding gene | Cytoband: 2q11.2 | ||
| Entrez ID: 7850 | HGNC ID: HGNC:5994 | Ensembl Gene: ENSG00000115590 | OMIM ID: 147811 |
| Drug and gene relationship at DGIdb | |||
IL1R2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa05166 | HTLV-I infection |
Expression of IL1R2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | IL1R2 | 7850 | 205403_at | -1.1593 | 0.4407 | |
| GSE20347 | IL1R2 | 7850 | 205403_at | -1.7806 | 0.0014 | |
| GSE23400 | IL1R2 | 7850 | 205403_at | -0.0669 | 0.8024 | |
| GSE26886 | IL1R2 | 7850 | 205403_at | -1.7833 | 0.0163 | |
| GSE29001 | IL1R2 | 7850 | 205403_at | -1.7672 | 0.0103 | |
| GSE38129 | IL1R2 | 7850 | 205403_at | -0.9792 | 0.0709 | |
| GSE45670 | IL1R2 | 7850 | 205403_at | -0.6083 | 0.2680 | |
| GSE53622 | IL1R2 | 7850 | 122711 | -0.7069 | 0.0135 | |
| GSE53624 | IL1R2 | 7850 | 122711 | -1.4839 | 0.0000 | |
| GSE63941 | IL1R2 | 7850 | 205403_at | 0.4216 | 0.6096 | |
| GSE77861 | IL1R2 | 7850 | 205403_at | -0.4493 | 0.6712 | |
| GSE97050 | IL1R2 | 7850 | A_24_P63019 | 0.0011 | 0.9986 | |
| SRP007169 | IL1R2 | 7850 | RNAseq | -1.8698 | 0.0155 | |
| SRP008496 | IL1R2 | 7850 | RNAseq | -1.6452 | 0.0249 | |
| SRP064894 | IL1R2 | 7850 | RNAseq | -0.6339 | 0.1993 | |
| SRP133303 | IL1R2 | 7850 | RNAseq | -1.1784 | 0.0008 | |
| SRP159526 | IL1R2 | 7850 | RNAseq | -2.1542 | 0.0039 | |
| SRP193095 | IL1R2 | 7850 | RNAseq | -0.6593 | 0.0717 | |
| SRP219564 | IL1R2 | 7850 | RNAseq | -2.9693 | 0.0004 | |
| TCGA | IL1R2 | 7850 | RNAseq | -0.0326 | 0.8998 |
Upregulated datasets: 0; Downregulated datasets: 9.
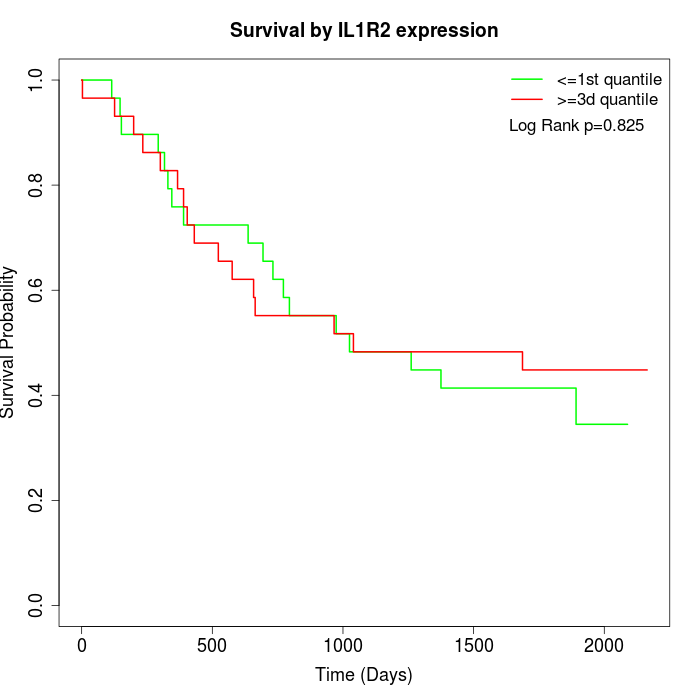
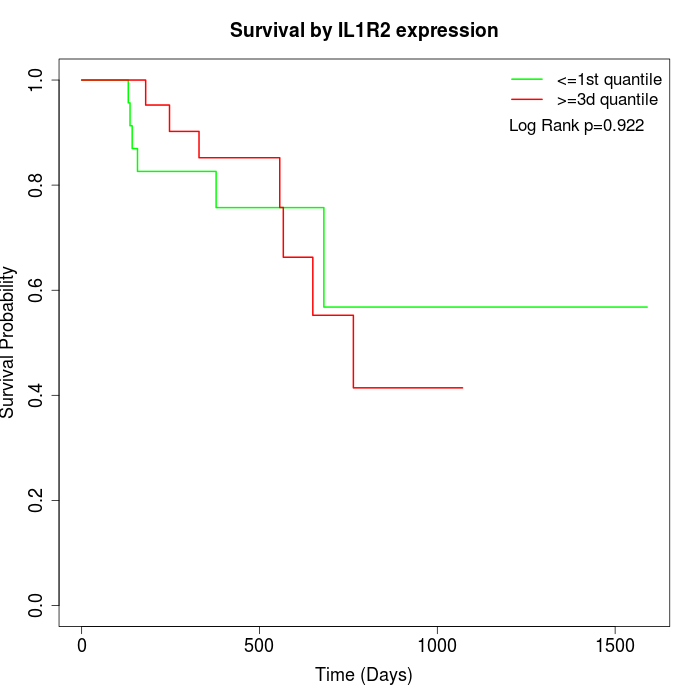
Survival by IL1R2 expression:
Note: Click image to view full size file.
Copy number change of IL1R2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | IL1R2 | 7850 | 5 | 2 | 23 | |
| GSE20123 | IL1R2 | 7850 | 5 | 2 | 23 | |
| GSE43470 | IL1R2 | 7850 | 5 | 1 | 37 | |
| GSE46452 | IL1R2 | 7850 | 1 | 4 | 54 | |
| GSE47630 | IL1R2 | 7850 | 7 | 0 | 33 | |
| GSE54993 | IL1R2 | 7850 | 0 | 6 | 64 | |
| GSE54994 | IL1R2 | 7850 | 11 | 0 | 42 | |
| GSE60625 | IL1R2 | 7850 | 0 | 3 | 8 | |
| GSE74703 | IL1R2 | 7850 | 5 | 1 | 30 | |
| GSE74704 | IL1R2 | 7850 | 3 | 1 | 16 | |
| TCGA | IL1R2 | 7850 | 34 | 6 | 56 |
Total number of gains: 76; Total number of losses: 26; Total Number of normals: 386.
Somatic mutations of IL1R2:
Generating mutation plots.
Highly correlated genes for IL1R2:
Showing top 20/318 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| IL1R2 | RAB18 | 0.728166 | 3 | 0 | 3 |
| IL1R2 | RNF144B | 0.682754 | 3 | 0 | 3 |
| IL1R2 | RSL1D1 | 0.673819 | 4 | 0 | 4 |
| IL1R2 | TMEM139 | 0.671082 | 3 | 0 | 3 |
| IL1R2 | PHYHD1 | 0.668815 | 3 | 0 | 3 |
| IL1R2 | EDEM1 | 0.664772 | 3 | 0 | 3 |
| IL1R2 | CRBN | 0.65791 | 3 | 0 | 3 |
| IL1R2 | WNT5A | 0.657024 | 6 | 0 | 5 |
| IL1R2 | PGM3 | 0.645652 | 3 | 0 | 3 |
| IL1R2 | AKR1C1 | 0.643214 | 3 | 0 | 3 |
| IL1R2 | FAM162B | 0.638739 | 4 | 0 | 3 |
| IL1R2 | LYSMD3 | 0.637985 | 4 | 0 | 4 |
| IL1R2 | TRERF1 | 0.632109 | 3 | 0 | 3 |
| IL1R2 | MTMR6 | 0.63024 | 4 | 0 | 3 |
| IL1R2 | CCDC65 | 0.625861 | 3 | 0 | 3 |
| IL1R2 | EXOC1 | 0.622475 | 4 | 0 | 3 |
| IL1R2 | HAUS1 | 0.621773 | 4 | 0 | 3 |
| IL1R2 | UBR2 | 0.620681 | 3 | 0 | 3 |
| IL1R2 | PDE4B | 0.620312 | 3 | 0 | 3 |
| IL1R2 | DDX1 | 0.619194 | 4 | 0 | 3 |
For details and further investigation, click here