| Full name: integrin alpha FG-GAP repeat containing 1 | Alias Symbol: CDA08|TIP|LNKN-1 | ||
| Type: protein-coding gene | Cytoband: 16q12.1 | ||
| Entrez ID: 81533 | HGNC ID: HGNC:30697 | Ensembl Gene: ENSG00000129636 | OMIM ID: 611803 |
| Drug and gene relationship at DGIdb | |||
Expression of ITFG1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ITFG1 | 81533 | 221449_s_at | -0.1734 | 0.8508 | |
| GSE20347 | ITFG1 | 81533 | 221449_s_at | 0.0242 | 0.9124 | |
| GSE23400 | ITFG1 | 81533 | 221449_s_at | 0.1353 | 0.0726 | |
| GSE26886 | ITFG1 | 81533 | 1556151_at | 0.8545 | 0.0000 | |
| GSE29001 | ITFG1 | 81533 | 221449_s_at | -0.0430 | 0.9139 | |
| GSE38129 | ITFG1 | 81533 | 221449_s_at | 0.1812 | 0.2545 | |
| GSE45670 | ITFG1 | 81533 | 221449_s_at | 0.1091 | 0.4600 | |
| GSE53622 | ITFG1 | 81533 | 146993 | 0.3700 | 0.0000 | |
| GSE53624 | ITFG1 | 81533 | 146993 | 0.1528 | 0.1103 | |
| GSE63941 | ITFG1 | 81533 | 1556151_at | -1.0549 | 0.0270 | |
| GSE77861 | ITFG1 | 81533 | 221449_s_at | 0.1203 | 0.6471 | |
| GSE97050 | ITFG1 | 81533 | A_23_P334123 | 0.4549 | 0.1187 | |
| SRP007169 | ITFG1 | 81533 | RNAseq | -0.1111 | 0.7438 | |
| SRP008496 | ITFG1 | 81533 | RNAseq | 0.0887 | 0.7423 | |
| SRP064894 | ITFG1 | 81533 | RNAseq | 0.0353 | 0.8327 | |
| SRP133303 | ITFG1 | 81533 | RNAseq | 0.5661 | 0.0000 | |
| SRP159526 | ITFG1 | 81533 | RNAseq | 0.4823 | 0.0928 | |
| SRP193095 | ITFG1 | 81533 | RNAseq | 0.0537 | 0.6766 | |
| SRP219564 | ITFG1 | 81533 | RNAseq | 0.0654 | 0.8899 | |
| TCGA | ITFG1 | 81533 | RNAseq | -0.0950 | 0.0473 |
Upregulated datasets: 0; Downregulated datasets: 1.
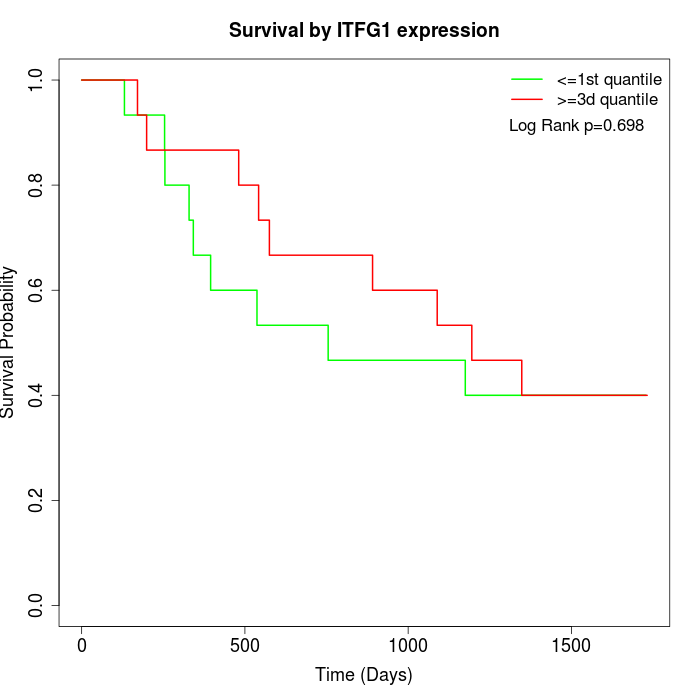
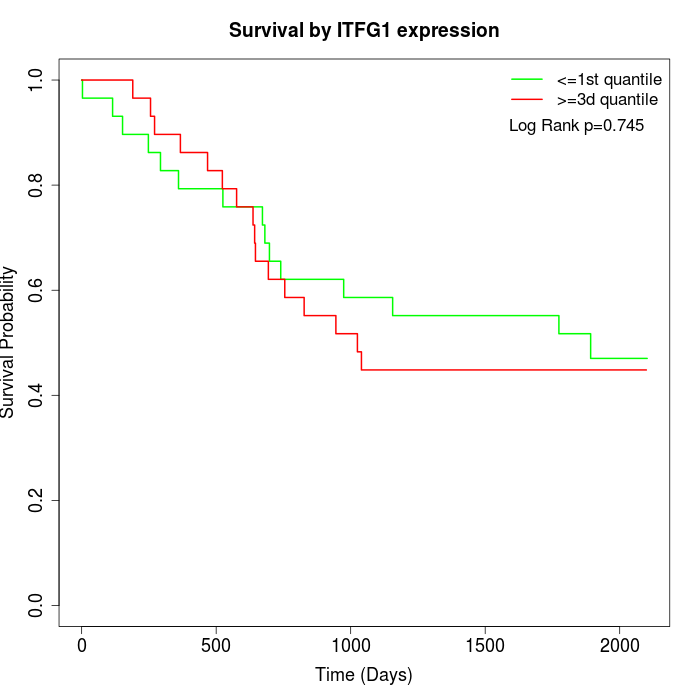
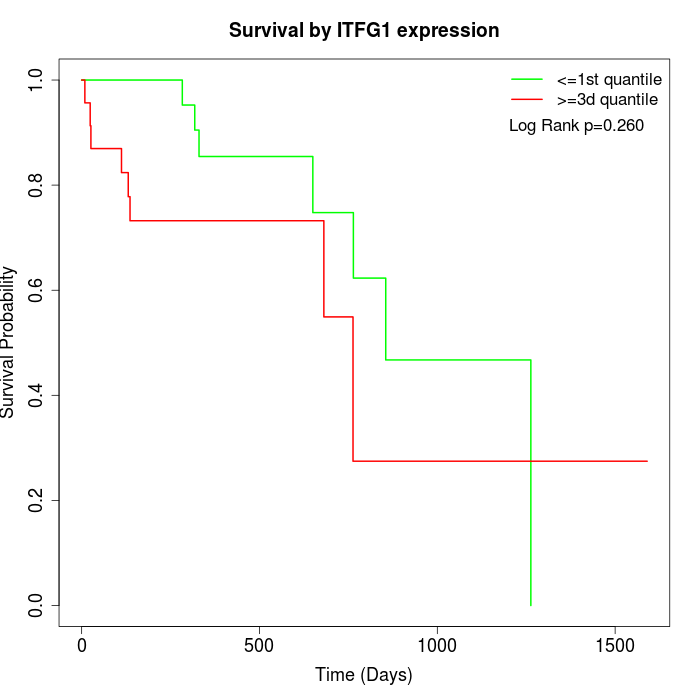
Survival by ITFG1 expression:
Note: Click image to view full size file.
Copy number change of ITFG1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ITFG1 | 81533 | 2 | 1 | 27 | |
| GSE20123 | ITFG1 | 81533 | 2 | 1 | 27 | |
| GSE43470 | ITFG1 | 81533 | 2 | 7 | 34 | |
| GSE46452 | ITFG1 | 81533 | 38 | 1 | 20 | |
| GSE47630 | ITFG1 | 81533 | 11 | 7 | 22 | |
| GSE54993 | ITFG1 | 81533 | 2 | 5 | 63 | |
| GSE54994 | ITFG1 | 81533 | 6 | 10 | 37 | |
| GSE60625 | ITFG1 | 81533 | 4 | 0 | 7 | |
| GSE74703 | ITFG1 | 81533 | 2 | 4 | 30 | |
| GSE74704 | ITFG1 | 81533 | 2 | 1 | 17 | |
| TCGA | ITFG1 | 81533 | 28 | 13 | 55 |
Total number of gains: 99; Total number of losses: 50; Total Number of normals: 339.
Somatic mutations of ITFG1:
Generating mutation plots.
Highly correlated genes for ITFG1:
Showing top 20/499 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ITFG1 | SRPX | 0.783884 | 3 | 0 | 3 |
| ITFG1 | DMXL2 | 0.757746 | 4 | 0 | 3 |
| ITFG1 | NUP50 | 0.745651 | 3 | 0 | 3 |
| ITFG1 | GLI3 | 0.736001 | 3 | 0 | 3 |
| ITFG1 | ZNF521 | 0.732625 | 3 | 0 | 3 |
| ITFG1 | BRAT1 | 0.731823 | 3 | 0 | 3 |
| ITFG1 | VPS16 | 0.730418 | 3 | 0 | 3 |
| ITFG1 | TMEM132A | 0.728441 | 3 | 0 | 3 |
| ITFG1 | NPAT | 0.726964 | 3 | 0 | 3 |
| ITFG1 | SLC2A3 | 0.724815 | 3 | 0 | 3 |
| ITFG1 | LMO1 | 0.72073 | 3 | 0 | 3 |
| ITFG1 | NFAT5 | 0.719044 | 3 | 0 | 3 |
| ITFG1 | ARHGAP31 | 0.715602 | 3 | 0 | 3 |
| ITFG1 | HAUS1 | 0.713785 | 4 | 0 | 3 |
| ITFG1 | SLC16A2 | 0.708129 | 4 | 0 | 4 |
| ITFG1 | TMEM14B | 0.705274 | 4 | 0 | 4 |
| ITFG1 | IGFBP5 | 0.702507 | 3 | 0 | 3 |
| ITFG1 | ARHGAP18 | 0.700202 | 4 | 0 | 4 |
| ITFG1 | LAPTM4B | 0.699599 | 4 | 0 | 3 |
| ITFG1 | MAPK9 | 0.694643 | 4 | 0 | 4 |
For details and further investigation, click here