| Full name: integrin subunit beta 1 binding protein 1 | Alias Symbol: ICAP-1A|ICAP-1B|ICAP1|ICAP1A|ICAP1B|ICAP-1alpha | ||
| Type: protein-coding gene | Cytoband: 2p25.1 | ||
| Entrez ID: 9270 | HGNC ID: HGNC:23927 | Ensembl Gene: ENSG00000119185 | OMIM ID: 607153 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of ITGB1BP1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ITGB1BP1 | 9270 | 203336_s_at | 0.1713 | 0.7455 | |
| GSE20347 | ITGB1BP1 | 9270 | 203336_s_at | 0.4660 | 0.0429 | |
| GSE23400 | ITGB1BP1 | 9270 | 203336_s_at | 0.5117 | 0.0000 | |
| GSE26886 | ITGB1BP1 | 9270 | 203336_s_at | 0.7921 | 0.0002 | |
| GSE29001 | ITGB1BP1 | 9270 | 203336_s_at | 0.4348 | 0.1393 | |
| GSE38129 | ITGB1BP1 | 9270 | 203336_s_at | 0.3583 | 0.0789 | |
| GSE45670 | ITGB1BP1 | 9270 | 203336_s_at | 0.1831 | 0.5562 | |
| GSE53622 | ITGB1BP1 | 9270 | 89190 | 0.5546 | 0.0000 | |
| GSE53624 | ITGB1BP1 | 9270 | 89190 | 0.7329 | 0.0000 | |
| GSE63941 | ITGB1BP1 | 9270 | 203336_s_at | -1.2590 | 0.0100 | |
| GSE77861 | ITGB1BP1 | 9270 | 203336_s_at | 0.2023 | 0.5659 | |
| GSE97050 | ITGB1BP1 | 9270 | A_23_P154507 | 0.5572 | 0.2289 | |
| SRP007169 | ITGB1BP1 | 9270 | RNAseq | 0.3434 | 0.3773 | |
| SRP008496 | ITGB1BP1 | 9270 | RNAseq | 0.6398 | 0.0257 | |
| SRP064894 | ITGB1BP1 | 9270 | RNAseq | 0.5000 | 0.0013 | |
| SRP133303 | ITGB1BP1 | 9270 | RNAseq | 0.4649 | 0.0001 | |
| SRP159526 | ITGB1BP1 | 9270 | RNAseq | 0.4710 | 0.0447 | |
| SRP193095 | ITGB1BP1 | 9270 | RNAseq | 0.3586 | 0.0060 | |
| SRP219564 | ITGB1BP1 | 9270 | RNAseq | -0.0441 | 0.9178 | |
| TCGA | ITGB1BP1 | 9270 | RNAseq | 0.1388 | 0.0084 |
Upregulated datasets: 0; Downregulated datasets: 1.
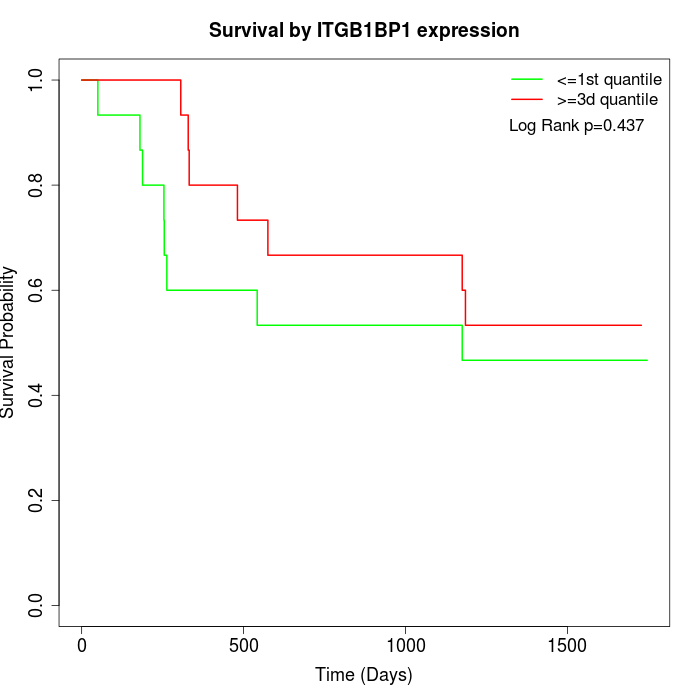
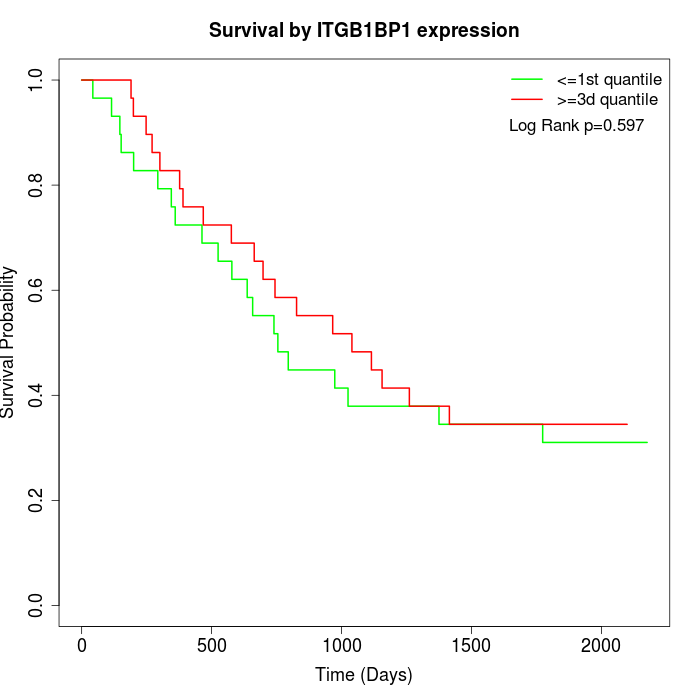
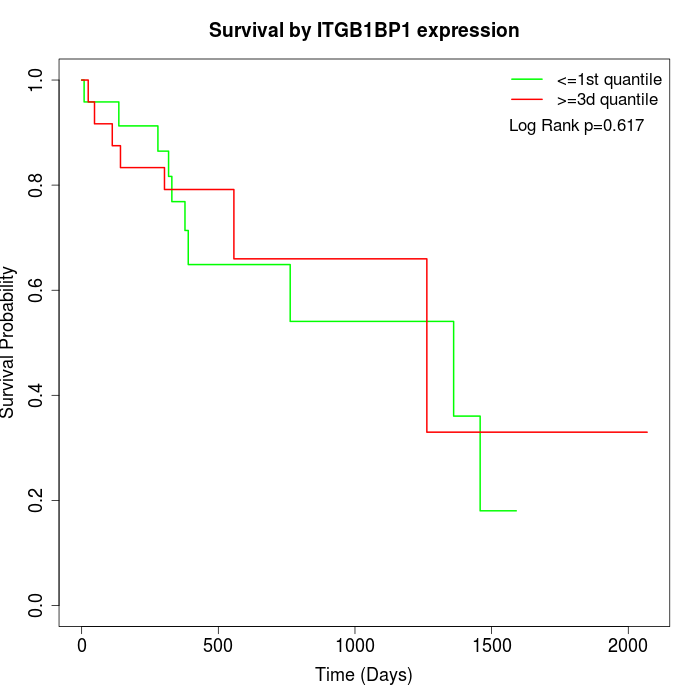
Survival by ITGB1BP1 expression:
Note: Click image to view full size file.
Copy number change of ITGB1BP1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ITGB1BP1 | 9270 | 11 | 2 | 17 | |
| GSE20123 | ITGB1BP1 | 9270 | 11 | 2 | 17 | |
| GSE43470 | ITGB1BP1 | 9270 | 2 | 1 | 40 | |
| GSE46452 | ITGB1BP1 | 9270 | 3 | 4 | 52 | |
| GSE47630 | ITGB1BP1 | 9270 | 7 | 0 | 33 | |
| GSE54993 | ITGB1BP1 | 9270 | 0 | 7 | 63 | |
| GSE54994 | ITGB1BP1 | 9270 | 11 | 1 | 41 | |
| GSE60625 | ITGB1BP1 | 9270 | 0 | 3 | 8 | |
| GSE74703 | ITGB1BP1 | 9270 | 2 | 0 | 34 | |
| GSE74704 | ITGB1BP1 | 9270 | 9 | 1 | 10 | |
| TCGA | ITGB1BP1 | 9270 | 34 | 6 | 56 |
Total number of gains: 90; Total number of losses: 27; Total Number of normals: 371.
Somatic mutations of ITGB1BP1:
Generating mutation plots.
Highly correlated genes for ITGB1BP1:
Showing top 20/751 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ITGB1BP1 | C6orf141 | 0.743274 | 3 | 0 | 3 |
| ITGB1BP1 | DCTN1 | 0.728269 | 3 | 0 | 3 |
| ITGB1BP1 | MCM3AP | 0.707995 | 3 | 0 | 3 |
| ITGB1BP1 | SARNP | 0.704492 | 3 | 0 | 3 |
| ITGB1BP1 | CHIC1 | 0.7037 | 3 | 0 | 3 |
| ITGB1BP1 | HNRNPUL1 | 0.69814 | 3 | 0 | 3 |
| ITGB1BP1 | THOP1 | 0.692211 | 3 | 0 | 3 |
| ITGB1BP1 | ZNF25 | 0.689935 | 3 | 0 | 3 |
| ITGB1BP1 | CEP250 | 0.685212 | 4 | 0 | 3 |
| ITGB1BP1 | RBBP6 | 0.684893 | 3 | 0 | 3 |
| ITGB1BP1 | FASTKD2 | 0.680877 | 3 | 0 | 3 |
| ITGB1BP1 | CD2BP2 | 0.680172 | 3 | 0 | 3 |
| ITGB1BP1 | C19orf54 | 0.67949 | 4 | 0 | 3 |
| ITGB1BP1 | CACHD1 | 0.678975 | 3 | 0 | 3 |
| ITGB1BP1 | USP32 | 0.677024 | 3 | 0 | 3 |
| ITGB1BP1 | GUSB | 0.674153 | 3 | 0 | 3 |
| ITGB1BP1 | VPS37A | 0.666953 | 3 | 0 | 3 |
| ITGB1BP1 | LETM2 | 0.663907 | 3 | 0 | 3 |
| ITGB1BP1 | LUC7L | 0.662412 | 3 | 0 | 3 |
| ITGB1BP1 | RAPGEF6 | 0.661704 | 3 | 0 | 3 |
For details and further investigation, click here