| Full name: potassium voltage-gated channel subfamily C member 2 | Alias Symbol: Kv3.2 | ||
| Type: protein-coding gene | Cytoband: 12q21.1 | ||
| Entrez ID: 3747 | HGNC ID: HGNC:6234 | Ensembl Gene: ENSG00000166006 | OMIM ID: 176256 |
| Drug and gene relationship at DGIdb | |||
Expression of KCNC2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNC2 | 3747 | 222289_at | -0.0143 | 0.9514 | |
| GSE20347 | KCNC2 | 3747 | 222289_at | -0.0469 | 0.5113 | |
| GSE23400 | KCNC2 | 3747 | 222289_at | -0.0495 | 0.0599 | |
| GSE26886 | KCNC2 | 3747 | 222289_at | 0.2683 | 0.0090 | |
| GSE29001 | KCNC2 | 3747 | 222289_at | 0.0055 | 0.9648 | |
| GSE38129 | KCNC2 | 3747 | 222289_at | -0.1165 | 0.0903 | |
| GSE45670 | KCNC2 | 3747 | 222289_at | -0.0950 | 0.2790 | |
| GSE53622 | KCNC2 | 3747 | 112718 | -0.0430 | 0.8238 | |
| GSE53624 | KCNC2 | 3747 | 112718 | 0.0462 | 0.7008 | |
| GSE63941 | KCNC2 | 3747 | 222289_at | 0.1911 | 0.1504 | |
| GSE77861 | KCNC2 | 3747 | 222289_at | 0.0069 | 0.9647 |
Upregulated datasets: 0; Downregulated datasets: 0.
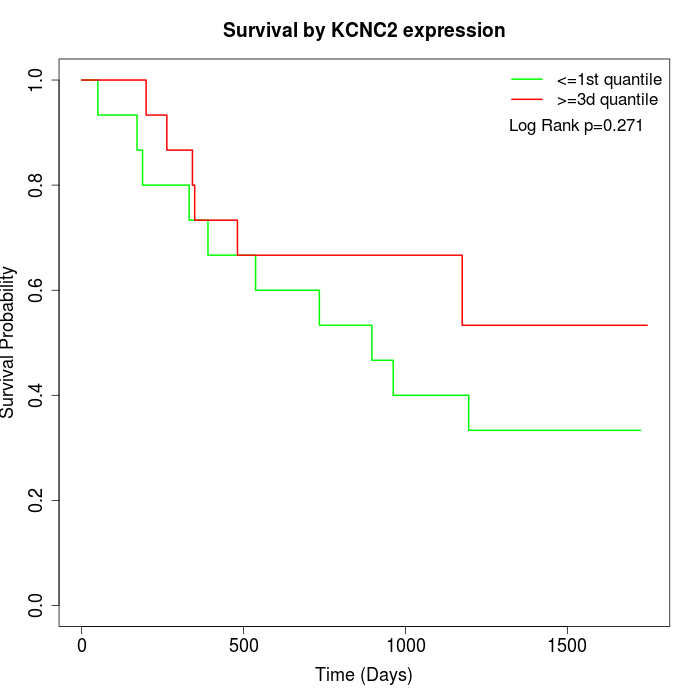
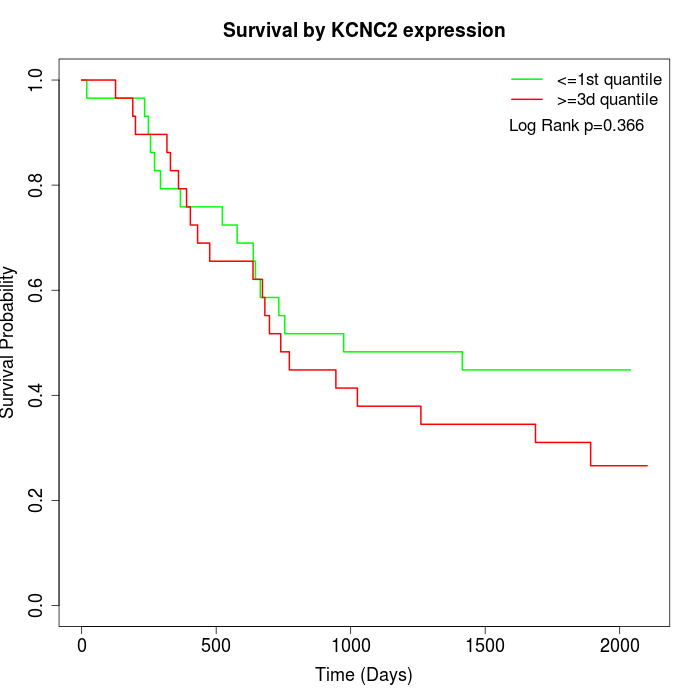
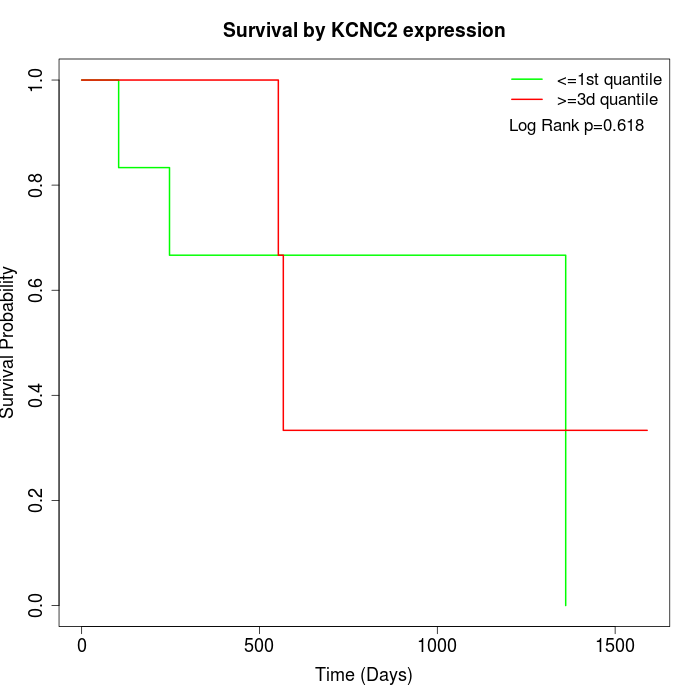
Survival by KCNC2 expression:
Note: Click image to view full size file.
Copy number change of KCNC2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNC2 | 3747 | 3 | 3 | 24 | |
| GSE20123 | KCNC2 | 3747 | 3 | 3 | 24 | |
| GSE43470 | KCNC2 | 3747 | 3 | 0 | 40 | |
| GSE46452 | KCNC2 | 3747 | 8 | 1 | 50 | |
| GSE47630 | KCNC2 | 3747 | 10 | 1 | 29 | |
| GSE54993 | KCNC2 | 3747 | 0 | 5 | 65 | |
| GSE54994 | KCNC2 | 3747 | 5 | 2 | 46 | |
| GSE60625 | KCNC2 | 3747 | 0 | 0 | 11 | |
| GSE74703 | KCNC2 | 3747 | 3 | 0 | 33 | |
| GSE74704 | KCNC2 | 3747 | 2 | 2 | 16 | |
| TCGA | KCNC2 | 3747 | 16 | 15 | 65 |
Total number of gains: 53; Total number of losses: 32; Total Number of normals: 403.
Somatic mutations of KCNC2:
Generating mutation plots.
Highly correlated genes for KCNC2:
Showing top 20/485 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNC2 | CACNA1H | 0.736427 | 3 | 0 | 3 |
| KCNC2 | SLC6A3 | 0.688887 | 3 | 0 | 3 |
| KCNC2 | LINGO1 | 0.685905 | 3 | 0 | 3 |
| KCNC2 | CPT1C | 0.683508 | 3 | 0 | 3 |
| KCNC2 | STK32C | 0.675247 | 3 | 0 | 3 |
| KCNC2 | HDAC7 | 0.672108 | 3 | 0 | 3 |
| KCNC2 | HDAC11 | 0.670792 | 3 | 0 | 3 |
| KCNC2 | LINC00598 | 0.667645 | 3 | 0 | 3 |
| KCNC2 | JSRP1 | 0.667173 | 4 | 0 | 3 |
| KCNC2 | SARDH | 0.665235 | 4 | 0 | 3 |
| KCNC2 | DSCAML1 | 0.660017 | 3 | 0 | 3 |
| KCNC2 | SELPLG | 0.657575 | 4 | 0 | 3 |
| KCNC2 | LTB4R2 | 0.655059 | 6 | 0 | 5 |
| KCNC2 | HGD | 0.65415 | 6 | 0 | 5 |
| KCNC2 | MYOG | 0.653903 | 7 | 0 | 7 |
| KCNC2 | KIF19 | 0.650485 | 3 | 0 | 3 |
| KCNC2 | TCF15 | 0.650297 | 5 | 0 | 4 |
| KCNC2 | PLD4 | 0.645085 | 3 | 0 | 3 |
| KCNC2 | SIX6 | 0.644714 | 6 | 0 | 5 |
| KCNC2 | IGFALS | 0.642397 | 8 | 0 | 6 |
For details and further investigation, click here