| Full name: DS cell adhesion molecule like 1 | Alias Symbol: KIAA1132 | ||
| Type: protein-coding gene | Cytoband: 11q23.3 | ||
| Entrez ID: 57453 | HGNC ID: HGNC:14656 | Ensembl Gene: ENSG00000177103 | OMIM ID: 611782 |
| Drug and gene relationship at DGIdb | |||
Expression of DSCAML1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DSCAML1 | 57453 | 234908_s_at | -0.1241 | 0.6865 | |
| GSE26886 | DSCAML1 | 57453 | 234908_s_at | 0.4495 | 0.0003 | |
| GSE45670 | DSCAML1 | 57453 | 234908_s_at | -0.0514 | 0.6764 | |
| GSE53622 | DSCAML1 | 57453 | 28704 | -0.8599 | 0.0000 | |
| GSE53624 | DSCAML1 | 57453 | 107419 | -1.6456 | 0.0000 | |
| GSE63941 | DSCAML1 | 57453 | 234908_s_at | 0.0962 | 0.6215 | |
| GSE77861 | DSCAML1 | 57453 | 234908_s_at | -0.1613 | 0.1422 | |
| SRP133303 | DSCAML1 | 57453 | RNAseq | -1.4831 | 0.0000 | |
| SRP159526 | DSCAML1 | 57453 | RNAseq | -2.0125 | 0.0046 | |
| TCGA | DSCAML1 | 57453 | RNAseq | -2.5263 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 4.
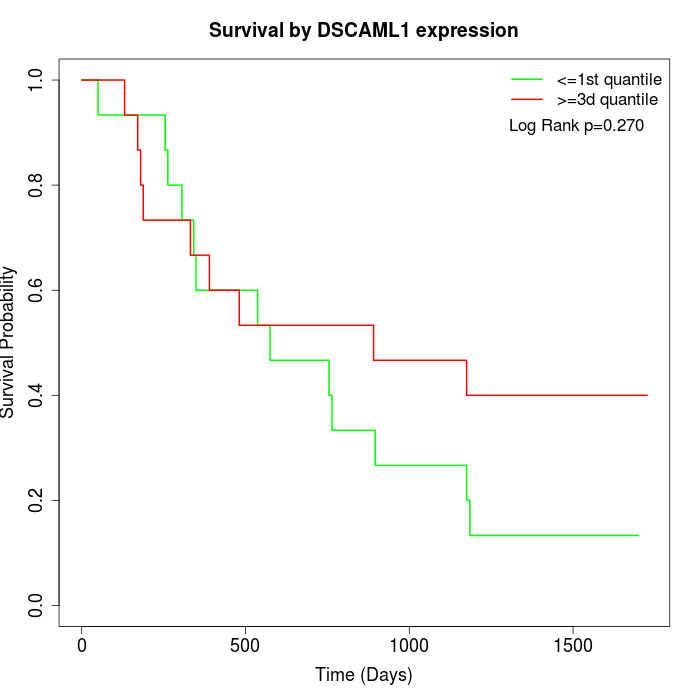
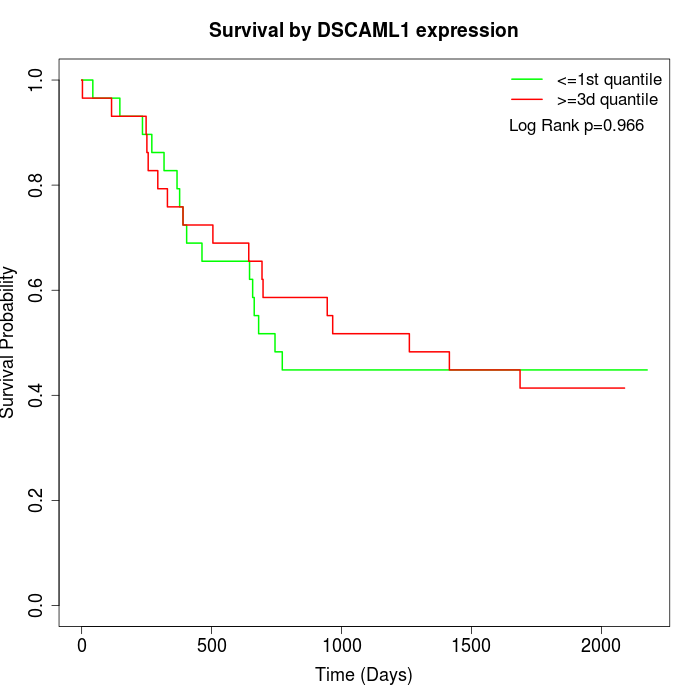
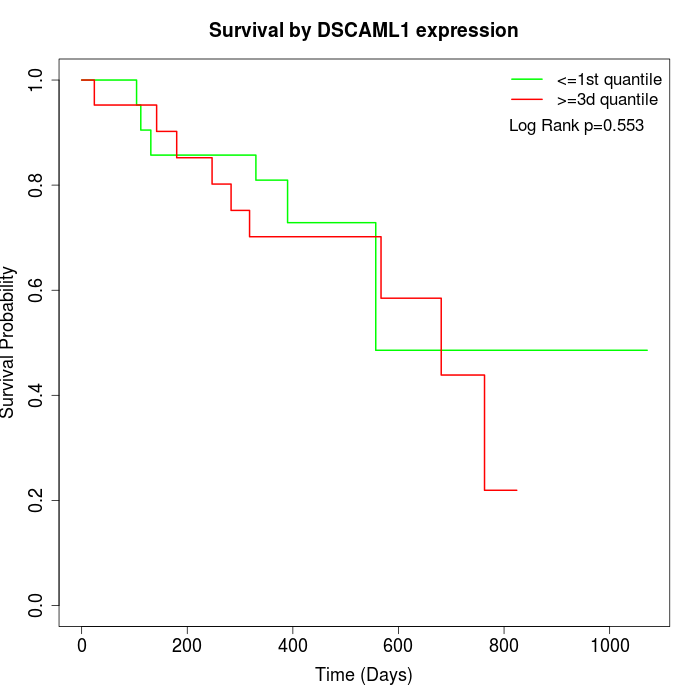
Survival by DSCAML1 expression:
Note: Click image to view full size file.
Copy number change of DSCAML1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DSCAML1 | 57453 | 1 | 13 | 16 | |
| GSE20123 | DSCAML1 | 57453 | 1 | 13 | 16 | |
| GSE43470 | DSCAML1 | 57453 | 2 | 9 | 32 | |
| GSE46452 | DSCAML1 | 57453 | 3 | 26 | 30 | |
| GSE47630 | DSCAML1 | 57453 | 2 | 19 | 19 | |
| GSE54993 | DSCAML1 | 57453 | 10 | 0 | 60 | |
| GSE54994 | DSCAML1 | 57453 | 5 | 19 | 29 | |
| GSE60625 | DSCAML1 | 57453 | 0 | 3 | 8 | |
| GSE74703 | DSCAML1 | 57453 | 1 | 6 | 29 | |
| GSE74704 | DSCAML1 | 57453 | 0 | 9 | 11 | |
| TCGA | DSCAML1 | 57453 | 5 | 50 | 41 |
Total number of gains: 30; Total number of losses: 167; Total Number of normals: 291.
Somatic mutations of DSCAML1:
Generating mutation plots.
Highly correlated genes for DSCAML1:
Showing top 20/165 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DSCAML1 | CNTFR | 0.671498 | 4 | 0 | 3 |
| DSCAML1 | KCNC2 | 0.660017 | 3 | 0 | 3 |
| DSCAML1 | TMEM151B | 0.657118 | 3 | 0 | 3 |
| DSCAML1 | PRRT2 | 0.649659 | 3 | 0 | 3 |
| DSCAML1 | IQSEC2 | 0.643707 | 3 | 0 | 3 |
| DSCAML1 | NLGN3 | 0.639215 | 5 | 0 | 5 |
| DSCAML1 | ELANE | 0.638177 | 3 | 0 | 3 |
| DSCAML1 | APOA4 | 0.631353 | 4 | 0 | 4 |
| DSCAML1 | GAL3ST1 | 0.631058 | 4 | 0 | 3 |
| DSCAML1 | HEYL | 0.628267 | 3 | 0 | 3 |
| DSCAML1 | TMEM255B | 0.6268 | 3 | 0 | 3 |
| DSCAML1 | SIGLEC8 | 0.624364 | 4 | 0 | 3 |
| DSCAML1 | FHL5 | 0.622444 | 3 | 0 | 3 |
| DSCAML1 | ITGA8 | 0.622346 | 4 | 0 | 3 |
| DSCAML1 | NR4A1 | 0.61873 | 3 | 0 | 3 |
| DSCAML1 | CRHR1 | 0.61715 | 3 | 0 | 3 |
| DSCAML1 | CMTM1 | 0.617005 | 3 | 0 | 3 |
| DSCAML1 | CDON | 0.614743 | 3 | 0 | 3 |
| DSCAML1 | FASLG | 0.612966 | 4 | 0 | 4 |
| DSCAML1 | EBF1 | 0.612777 | 3 | 0 | 3 |
For details and further investigation, click here