| Full name: potassium inwardly rectifying channel subfamily J member 15 | Alias Symbol: Kir4.2|Kir1.3|IRKK | ||
| Type: protein-coding gene | Cytoband: 21q22.13-q22.2 | ||
| Entrez ID: 3772 | HGNC ID: HGNC:6261 | Ensembl Gene: ENSG00000157551 | OMIM ID: 602106 |
| Drug and gene relationship at DGIdb | |||
KCNJ15 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04971 | Gastric acid secretion |
Expression of KCNJ15:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNJ15 | 3772 | 210119_at | 0.4813 | 0.6806 | |
| GSE20347 | KCNJ15 | 3772 | 210119_at | -0.2155 | 0.6068 | |
| GSE23400 | KCNJ15 | 3772 | 211806_s_at | 0.0902 | 0.1485 | |
| GSE26886 | KCNJ15 | 3772 | 210119_at | 0.2495 | 0.6986 | |
| GSE29001 | KCNJ15 | 3772 | 210119_at | 0.1381 | 0.7966 | |
| GSE38129 | KCNJ15 | 3772 | 210119_at | 0.0229 | 0.9633 | |
| GSE45670 | KCNJ15 | 3772 | 210119_at | 0.7561 | 0.0470 | |
| GSE53622 | KCNJ15 | 3772 | 70720 | 0.2184 | 0.3676 | |
| GSE53624 | KCNJ15 | 3772 | 70720 | 0.3611 | 0.0272 | |
| GSE63941 | KCNJ15 | 3772 | 211806_s_at | 1.4176 | 0.2214 | |
| GSE77861 | KCNJ15 | 3772 | 210119_at | 0.0170 | 0.9802 | |
| GSE97050 | KCNJ15 | 3772 | A_33_P3267532 | 0.4116 | 0.5590 | |
| SRP007169 | KCNJ15 | 3772 | RNAseq | -1.3110 | 0.0119 | |
| SRP008496 | KCNJ15 | 3772 | RNAseq | -0.3976 | 0.5235 | |
| SRP064894 | KCNJ15 | 3772 | RNAseq | 0.4533 | 0.1163 | |
| SRP133303 | KCNJ15 | 3772 | RNAseq | -0.3175 | 0.2151 | |
| SRP159526 | KCNJ15 | 3772 | RNAseq | 0.0650 | 0.9140 | |
| SRP193095 | KCNJ15 | 3772 | RNAseq | -0.3121 | 0.1896 | |
| SRP219564 | KCNJ15 | 3772 | RNAseq | 0.5909 | 0.4852 | |
| TCGA | KCNJ15 | 3772 | RNAseq | -0.0746 | 0.7118 |
Upregulated datasets: 0; Downregulated datasets: 1.
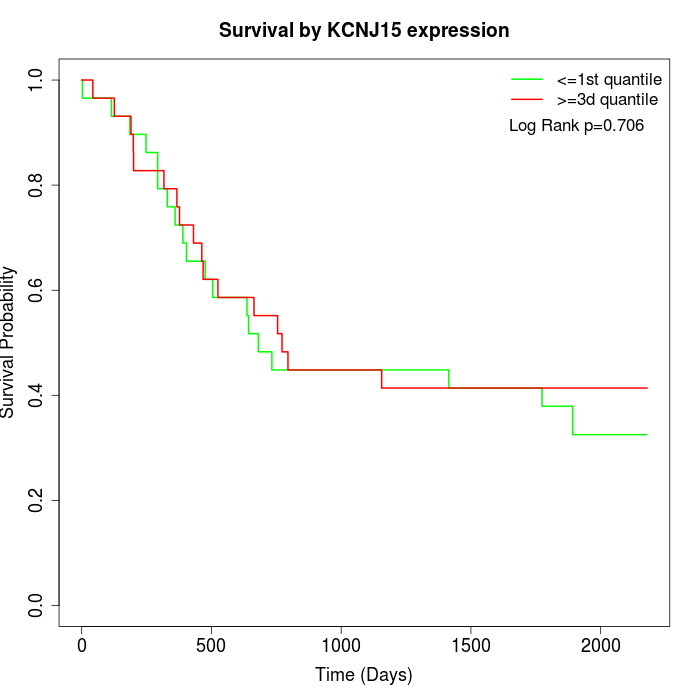
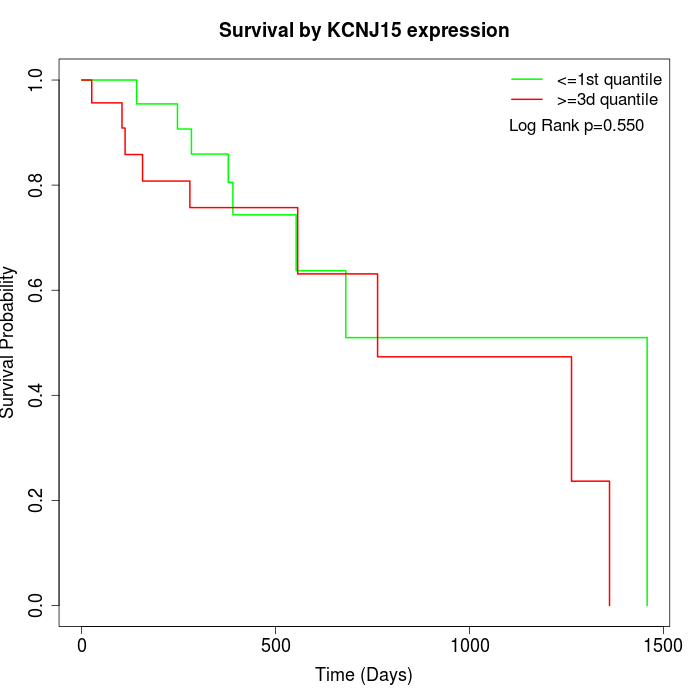
Survival by KCNJ15 expression:
Note: Click image to view full size file.
Copy number change of KCNJ15:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNJ15 | 3772 | 2 | 9 | 19 | |
| GSE20123 | KCNJ15 | 3772 | 2 | 10 | 18 | |
| GSE43470 | KCNJ15 | 3772 | 1 | 11 | 31 | |
| GSE46452 | KCNJ15 | 3772 | 1 | 21 | 37 | |
| GSE47630 | KCNJ15 | 3772 | 6 | 17 | 17 | |
| GSE54993 | KCNJ15 | 3772 | 8 | 1 | 61 | |
| GSE54994 | KCNJ15 | 3772 | 3 | 9 | 41 | |
| GSE60625 | KCNJ15 | 3772 | 0 | 0 | 11 | |
| GSE74703 | KCNJ15 | 3772 | 1 | 8 | 27 | |
| GSE74704 | KCNJ15 | 3772 | 1 | 7 | 12 | |
| TCGA | KCNJ15 | 3772 | 8 | 41 | 47 |
Total number of gains: 33; Total number of losses: 134; Total Number of normals: 321.
Somatic mutations of KCNJ15:
Generating mutation plots.
Highly correlated genes for KCNJ15:
Showing top 20/167 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNJ15 | CDC27 | 0.705829 | 3 | 0 | 3 |
| KCNJ15 | PNO1 | 0.705817 | 3 | 0 | 3 |
| KCNJ15 | SUPT16H | 0.682119 | 3 | 0 | 3 |
| KCNJ15 | SERINC2 | 0.681694 | 3 | 0 | 3 |
| KCNJ15 | AMD1 | 0.678788 | 3 | 0 | 3 |
| KCNJ15 | UBLCP1 | 0.676498 | 4 | 0 | 3 |
| KCNJ15 | EED | 0.666741 | 3 | 0 | 3 |
| KCNJ15 | TMCO6 | 0.659702 | 3 | 0 | 3 |
| KCNJ15 | FAM83G | 0.658661 | 4 | 0 | 4 |
| KCNJ15 | SPECC1 | 0.658565 | 3 | 0 | 3 |
| KCNJ15 | MRPS27 | 0.658508 | 3 | 0 | 3 |
| KCNJ15 | ASCC3 | 0.657371 | 5 | 0 | 3 |
| KCNJ15 | AIMP1 | 0.656977 | 3 | 0 | 3 |
| KCNJ15 | TRADD | 0.656093 | 4 | 0 | 4 |
| KCNJ15 | SNX14 | 0.653777 | 3 | 0 | 3 |
| KCNJ15 | TOX4 | 0.646448 | 4 | 0 | 4 |
| KCNJ15 | N4BP1 | 0.644836 | 3 | 0 | 3 |
| KCNJ15 | SCAMP1 | 0.644251 | 4 | 0 | 3 |
| KCNJ15 | CSTF3 | 0.642606 | 3 | 0 | 3 |
| KCNJ15 | AMPD3 | 0.641066 | 4 | 0 | 3 |
For details and further investigation, click here