| Full name: cell division cycle 27 | Alias Symbol: APC3|ANAPC3|NUC2 | ||
| Type: protein-coding gene | Cytoband: 17q21.32 | ||
| Entrez ID: 996 | HGNC ID: HGNC:1728 | Ensembl Gene: ENSG00000004897 | OMIM ID: 116946 |
| Drug and gene relationship at DGIdb | |||
CDC27 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04110 | Cell cycle | |
| hsa04114 | Oocyte meiosis | |
| hsa04914 | Progesterone-mediated oocyte maturation | |
| hsa05166 | HTLV-I infection |
Expression of CDC27:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CDC27 | 996 | 217880_at | 0.2371 | 0.4716 | |
| GSE20347 | CDC27 | 996 | 217880_at | 0.1592 | 0.2078 | |
| GSE23400 | CDC27 | 996 | 217878_s_at | 0.1222 | 0.1448 | |
| GSE26886 | CDC27 | 996 | 217880_at | 0.1712 | 0.4150 | |
| GSE29001 | CDC27 | 996 | 217879_at | 0.4714 | 0.0500 | |
| GSE38129 | CDC27 | 996 | 217880_at | 0.2967 | 0.0051 | |
| GSE45670 | CDC27 | 996 | 217878_s_at | 0.0337 | 0.9035 | |
| GSE53622 | CDC27 | 996 | 107997 | 0.5212 | 0.0000 | |
| GSE53624 | CDC27 | 996 | 107997 | 0.5620 | 0.0000 | |
| GSE63941 | CDC27 | 996 | 217880_at | -0.9624 | 0.0290 | |
| GSE77861 | CDC27 | 996 | 217879_at | 0.6419 | 0.0283 | |
| GSE97050 | CDC27 | 996 | A_23_P66777 | 0.4511 | 0.1858 | |
| SRP007169 | CDC27 | 996 | RNAseq | 0.6333 | 0.0449 | |
| SRP008496 | CDC27 | 996 | RNAseq | 0.6231 | 0.0002 | |
| SRP064894 | CDC27 | 996 | RNAseq | 0.5724 | 0.0008 | |
| SRP133303 | CDC27 | 996 | RNAseq | 0.5478 | 0.0001 | |
| SRP159526 | CDC27 | 996 | RNAseq | 0.3294 | 0.1345 | |
| SRP193095 | CDC27 | 996 | RNAseq | 0.3033 | 0.0019 | |
| SRP219564 | CDC27 | 996 | RNAseq | 0.3993 | 0.1328 | |
| TCGA | CDC27 | 996 | RNAseq | 0.1195 | 0.0101 |
Upregulated datasets: 0; Downregulated datasets: 0.
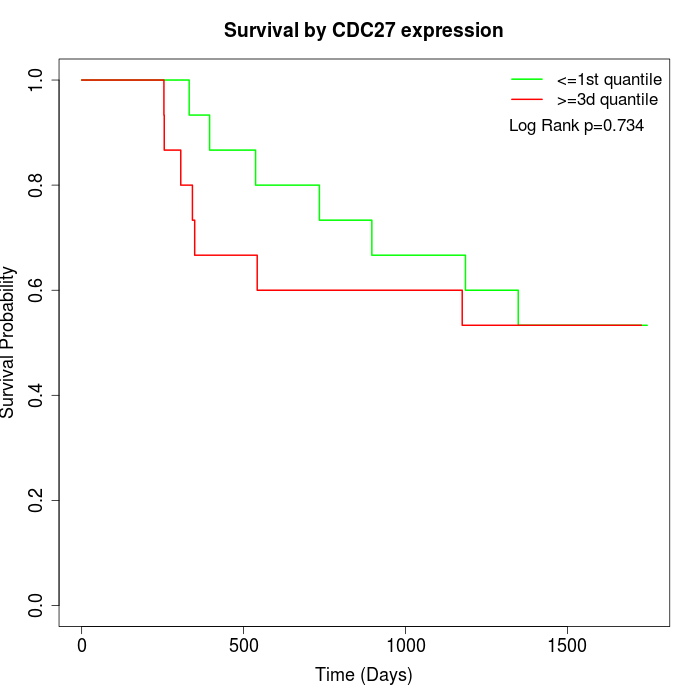
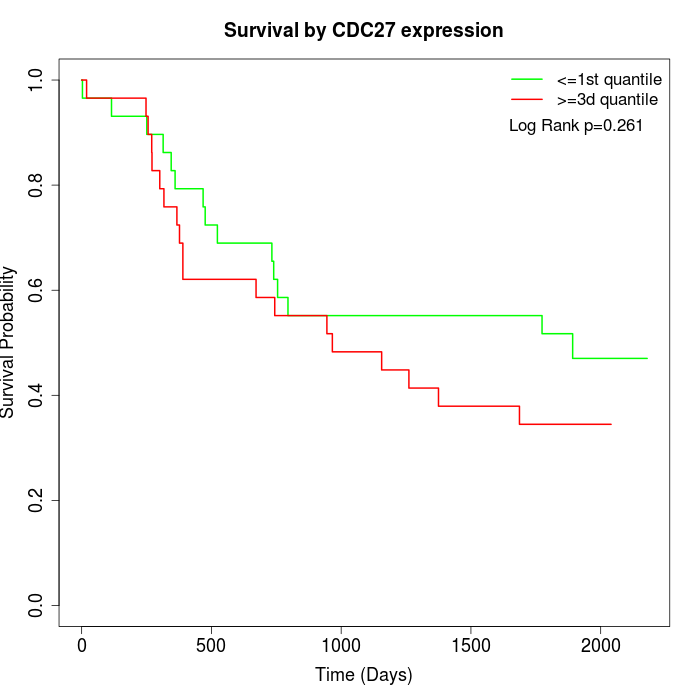
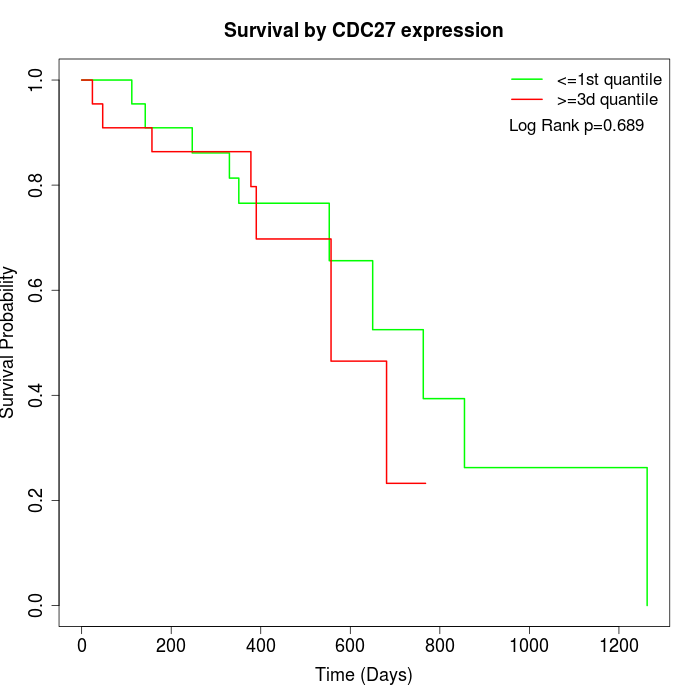
Survival by CDC27 expression:
Note: Click image to view full size file.
Copy number change of CDC27:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CDC27 | 996 | 5 | 2 | 23 | |
| GSE20123 | CDC27 | 996 | 5 | 2 | 23 | |
| GSE43470 | CDC27 | 996 | 1 | 2 | 40 | |
| GSE46452 | CDC27 | 996 | 32 | 0 | 27 | |
| GSE47630 | CDC27 | 996 | 9 | 0 | 31 | |
| GSE54993 | CDC27 | 996 | 2 | 4 | 64 | |
| GSE54994 | CDC27 | 996 | 11 | 5 | 37 | |
| GSE60625 | CDC27 | 996 | 4 | 0 | 7 | |
| GSE74703 | CDC27 | 996 | 1 | 1 | 34 | |
| GSE74704 | CDC27 | 996 | 4 | 1 | 15 | |
| TCGA | CDC27 | 996 | 26 | 6 | 64 |
Total number of gains: 100; Total number of losses: 23; Total Number of normals: 365.
Somatic mutations of CDC27:
Generating mutation plots.
Highly correlated genes for CDC27:
Showing top 20/1026 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CDC27 | UBA6 | 0.807975 | 3 | 0 | 3 |
| CDC27 | PLEKHB2 | 0.79494 | 3 | 0 | 3 |
| CDC27 | NF1 | 0.762184 | 3 | 0 | 3 |
| CDC27 | ATP1A1 | 0.748185 | 3 | 0 | 3 |
| CDC27 | TES | 0.734677 | 3 | 0 | 3 |
| CDC27 | RAB34 | 0.732944 | 3 | 0 | 3 |
| CDC27 | CDC37 | 0.730546 | 3 | 0 | 3 |
| CDC27 | PLEKHO2 | 0.727265 | 3 | 0 | 3 |
| CDC27 | KHDC1 | 0.721768 | 3 | 0 | 3 |
| CDC27 | PYCR2 | 0.721708 | 3 | 0 | 3 |
| CDC27 | HOOK3 | 0.721686 | 3 | 0 | 3 |
| CDC27 | WDR36 | 0.716371 | 4 | 0 | 4 |
| CDC27 | PPIP5K2 | 0.713233 | 3 | 0 | 3 |
| CDC27 | SCNM1 | 0.711801 | 3 | 0 | 3 |
| CDC27 | FADS1 | 0.708225 | 3 | 0 | 3 |
| CDC27 | SPOPL | 0.707651 | 3 | 0 | 3 |
| CDC27 | PPIG | 0.707199 | 4 | 0 | 3 |
| CDC27 | SASS6 | 0.706818 | 4 | 0 | 3 |
| CDC27 | KCNJ15 | 0.705829 | 3 | 0 | 3 |
| CDC27 | RFTN1 | 0.70247 | 4 | 0 | 3 |
For details and further investigation, click here