| Full name: potassium two pore domain channel subfamily K member 12 | Alias Symbol: THIK-2|THIK2|K2p12.1 | ||
| Type: protein-coding gene | Cytoband: 2p16.3 | ||
| Entrez ID: 56660 | HGNC ID: HGNC:6274 | Ensembl Gene: ENSG00000184261 | OMIM ID: 607366 |
| Drug and gene relationship at DGIdb | |||
Expression of KCNK12:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNK12 | 56660 | 220448_at | -0.1910 | 0.5137 | |
| GSE20347 | KCNK12 | 56660 | 220448_at | -0.0753 | 0.3046 | |
| GSE23400 | KCNK12 | 56660 | 220448_at | -0.0801 | 0.0009 | |
| GSE26886 | KCNK12 | 56660 | 220448_at | -0.0811 | 0.5162 | |
| GSE29001 | KCNK12 | 56660 | 220448_at | -0.2422 | 0.1279 | |
| GSE38129 | KCNK12 | 56660 | 220448_at | -0.0845 | 0.4813 | |
| GSE45670 | KCNK12 | 56660 | 220448_at | -0.0120 | 0.9401 | |
| GSE53622 | KCNK12 | 56660 | 126550 | 0.5312 | 0.0000 | |
| GSE53624 | KCNK12 | 56660 | 126550 | -0.2237 | 0.0153 | |
| GSE63941 | KCNK12 | 56660 | 220448_at | -4.5112 | 0.0000 | |
| GSE77861 | KCNK12 | 56660 | 220448_at | -0.1526 | 0.1623 | |
| GSE97050 | KCNK12 | 56660 | A_33_P3256510 | 0.0835 | 0.6773 | |
| SRP007169 | KCNK12 | 56660 | RNAseq | 0.4848 | 0.5820 | |
| SRP133303 | KCNK12 | 56660 | RNAseq | -0.1501 | 0.4942 | |
| SRP159526 | KCNK12 | 56660 | RNAseq | 0.9809 | 0.1167 | |
| SRP193095 | KCNK12 | 56660 | RNAseq | 0.7617 | 0.0001 | |
| SRP219564 | KCNK12 | 56660 | RNAseq | 0.1188 | 0.7737 | |
| TCGA | KCNK12 | 56660 | RNAseq | 0.1389 | 0.7652 |
Upregulated datasets: 0; Downregulated datasets: 1.
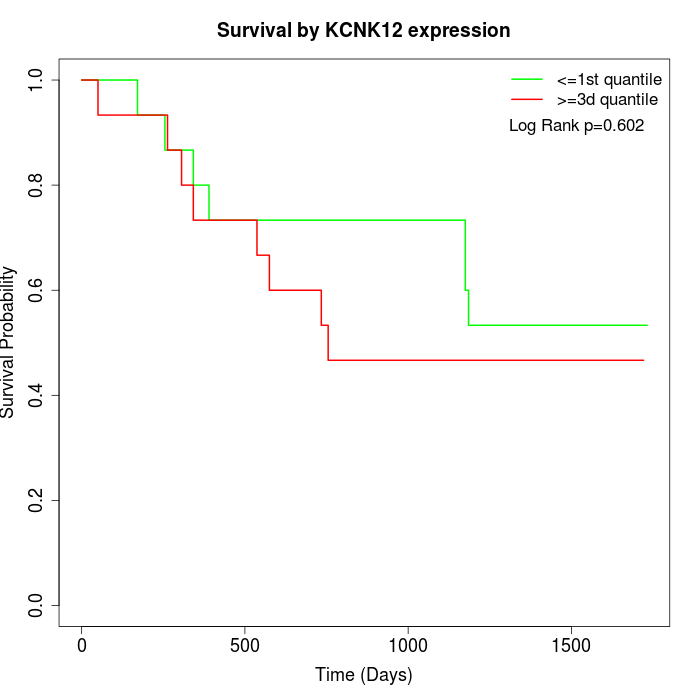
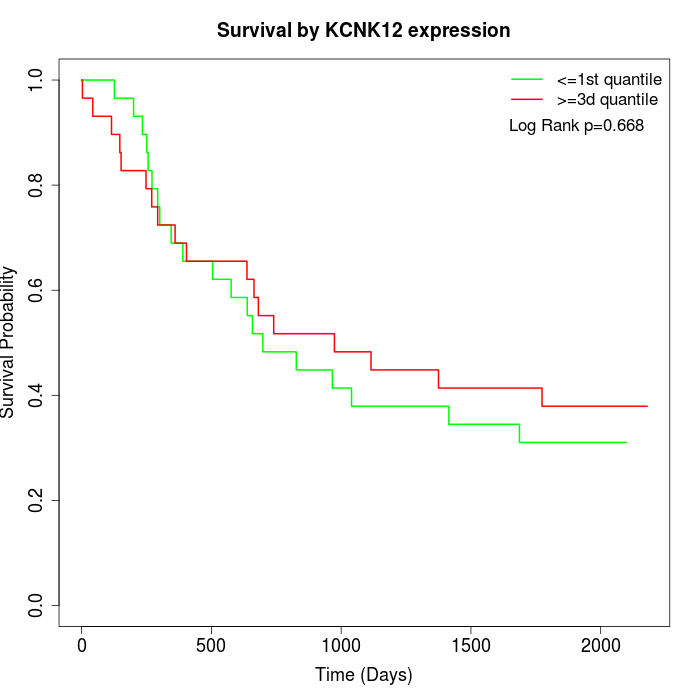
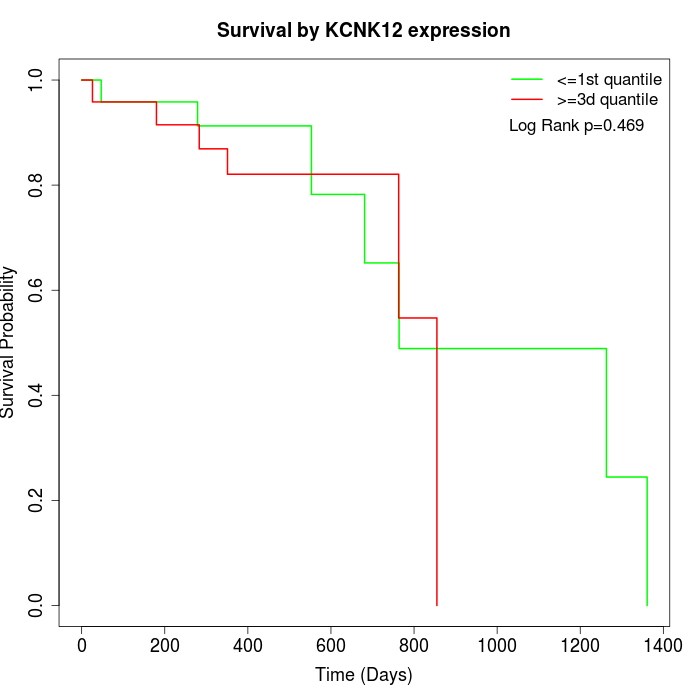
Survival by KCNK12 expression:
Note: Click image to view full size file.
Copy number change of KCNK12:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNK12 | 56660 | 8 | 1 | 21 | |
| GSE20123 | KCNK12 | 56660 | 7 | 1 | 22 | |
| GSE43470 | KCNK12 | 56660 | 5 | 0 | 38 | |
| GSE46452 | KCNK12 | 56660 | 1 | 4 | 54 | |
| GSE47630 | KCNK12 | 56660 | 8 | 0 | 32 | |
| GSE54993 | KCNK12 | 56660 | 0 | 5 | 65 | |
| GSE54994 | KCNK12 | 56660 | 12 | 0 | 41 | |
| GSE60625 | KCNK12 | 56660 | 0 | 3 | 8 | |
| GSE74703 | KCNK12 | 56660 | 5 | 0 | 31 | |
| GSE74704 | KCNK12 | 56660 | 8 | 1 | 11 | |
| TCGA | KCNK12 | 56660 | 36 | 2 | 58 |
Total number of gains: 90; Total number of losses: 17; Total Number of normals: 381.
Somatic mutations of KCNK12:
Generating mutation plots.
Highly correlated genes for KCNK12:
Showing top 20/75 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNK12 | SVOPL | 0.765904 | 3 | 0 | 3 |
| KCNK12 | NICN1 | 0.719489 | 3 | 0 | 3 |
| KCNK12 | NXF3 | 0.704089 | 4 | 0 | 4 |
| KCNK12 | ABCC6 | 0.696493 | 3 | 0 | 3 |
| KCNK12 | D2HGDH | 0.693489 | 4 | 0 | 3 |
| KCNK12 | CCL13 | 0.683003 | 4 | 0 | 3 |
| KCNK12 | BBOX1 | 0.669471 | 3 | 0 | 3 |
| KCNK12 | MAP3K6 | 0.666326 | 3 | 0 | 3 |
| KCNK12 | SCGN | 0.661405 | 4 | 0 | 4 |
| KCNK12 | ZDHHC15 | 0.658096 | 3 | 0 | 3 |
| KCNK12 | C8orf48 | 0.65499 | 3 | 0 | 3 |
| KCNK12 | SCN3A | 0.640094 | 5 | 0 | 3 |
| KCNK12 | SYPL2 | 0.638596 | 4 | 0 | 3 |
| KCNK12 | CDHR5 | 0.637455 | 5 | 0 | 4 |
| KCNK12 | ADRA2A | 0.63702 | 4 | 0 | 3 |
| KCNK12 | LRRC2 | 0.637001 | 4 | 0 | 3 |
| KCNK12 | FANK1 | 0.632125 | 5 | 0 | 3 |
| KCNK12 | TMEM217 | 0.629907 | 6 | 0 | 5 |
| KCNK12 | RAET1E | 0.629638 | 3 | 0 | 3 |
| KCNK12 | DAB1 | 0.619704 | 5 | 0 | 3 |
For details and further investigation, click here