| Full name: potassium voltage-gated channel subfamily Q member 1 | Alias Symbol: Kv7.1|KCNA8|KVLQT1|JLNS1|LQT1 | ||
| Type: protein-coding gene | Cytoband: 11p15.5-p15.4 | ||
| Entrez ID: 3784 | HGNC ID: HGNC:6294 | Ensembl Gene: ENSG00000053918 | OMIM ID: 607542 |
| Drug and gene relationship at DGIdb | |||
KCNQ1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04261 | Adrenergic signaling in cardiomyocytes | |
| hsa04971 | Gastric acid secretion | |
| hsa04972 | Pancreatic secretion | |
| hsa05110 | Vibrio cholerae infection |
Expression of KCNQ1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNQ1 | 3784 | 211217_s_at | 0.1166 | 0.5845 | |
| GSE20347 | KCNQ1 | 3784 | 211217_s_at | -0.0890 | 0.3804 | |
| GSE23400 | KCNQ1 | 3784 | 211217_s_at | -0.1071 | 0.0016 | |
| GSE26886 | KCNQ1 | 3784 | 211217_s_at | -0.0107 | 0.9414 | |
| GSE29001 | KCNQ1 | 3784 | 211217_s_at | -0.2307 | 0.0809 | |
| GSE38129 | KCNQ1 | 3784 | 211217_s_at | -0.2069 | 0.0976 | |
| GSE45670 | KCNQ1 | 3784 | 211217_s_at | 0.0610 | 0.5547 | |
| GSE53622 | KCNQ1 | 3784 | 89705 | 0.0909 | 0.0951 | |
| GSE53624 | KCNQ1 | 3784 | 89705 | 0.1297 | 0.0304 | |
| GSE63941 | KCNQ1 | 3784 | 211217_s_at | 0.4561 | 0.0146 | |
| GSE77861 | KCNQ1 | 3784 | 211217_s_at | -0.1168 | 0.3343 | |
| GSE97050 | KCNQ1 | 3784 | A_23_P429977 | 0.2210 | 0.3138 | |
| SRP064894 | KCNQ1 | 3784 | RNAseq | 0.3865 | 0.2690 | |
| SRP133303 | KCNQ1 | 3784 | RNAseq | -0.0140 | 0.9448 | |
| SRP159526 | KCNQ1 | 3784 | RNAseq | -0.1618 | 0.7023 | |
| SRP193095 | KCNQ1 | 3784 | RNAseq | -0.2189 | 0.6701 | |
| SRP219564 | KCNQ1 | 3784 | RNAseq | 2.3298 | 0.0017 | |
| TCGA | KCNQ1 | 3784 | RNAseq | -1.2576 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 1.
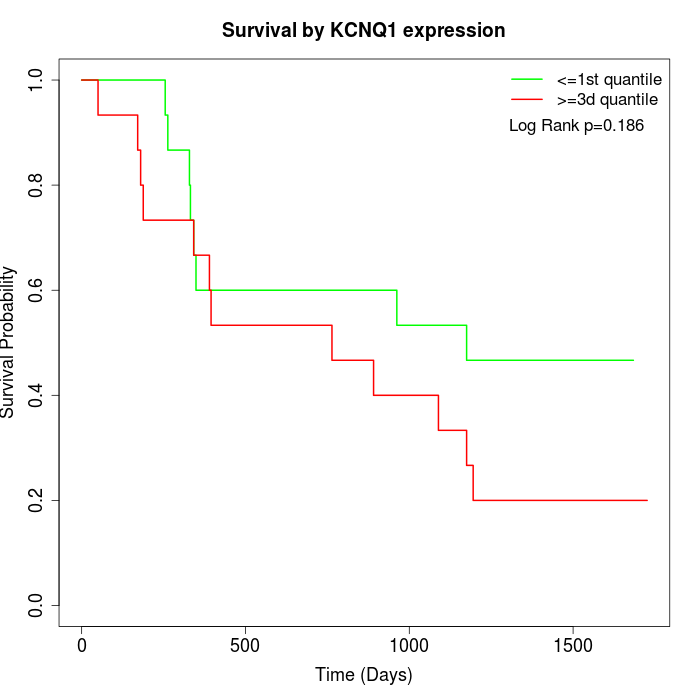
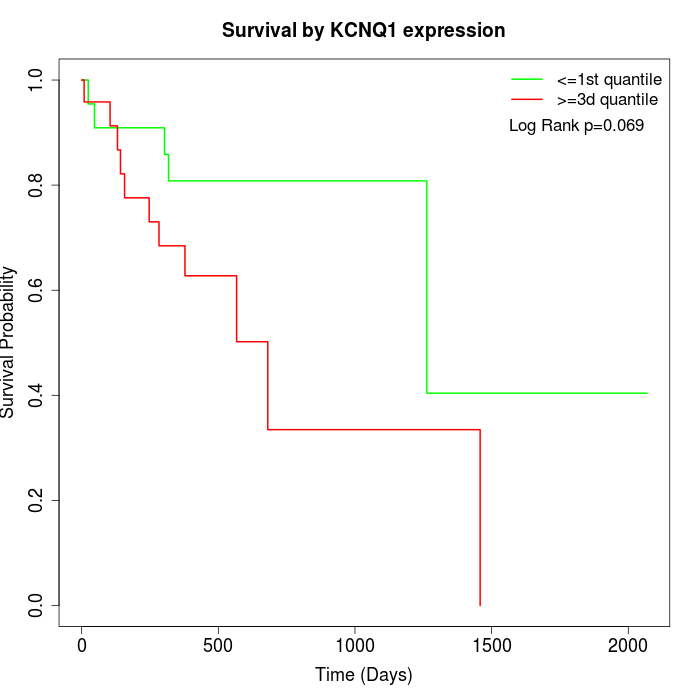
Survival by KCNQ1 expression:
Note: Click image to view full size file.
Copy number change of KCNQ1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNQ1 | 3784 | 1 | 11 | 18 | |
| GSE20123 | KCNQ1 | 3784 | 1 | 12 | 17 | |
| GSE43470 | KCNQ1 | 3784 | 1 | 10 | 32 | |
| GSE46452 | KCNQ1 | 3784 | 7 | 8 | 44 | |
| GSE47630 | KCNQ1 | 3784 | 4 | 12 | 24 | |
| GSE54993 | KCNQ1 | 3784 | 3 | 1 | 66 | |
| GSE54994 | KCNQ1 | 3784 | 1 | 12 | 40 | |
| GSE60625 | KCNQ1 | 3784 | 0 | 0 | 11 | |
| GSE74703 | KCNQ1 | 3784 | 1 | 8 | 27 | |
| GSE74704 | KCNQ1 | 3784 | 0 | 8 | 12 | |
| TCGA | KCNQ1 | 3784 | 8 | 39 | 49 |
Total number of gains: 27; Total number of losses: 121; Total Number of normals: 340.
Somatic mutations of KCNQ1:
Generating mutation plots.
Highly correlated genes for KCNQ1:
Showing top 20/687 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNQ1 | RAPGEF5 | 0.669997 | 3 | 0 | 3 |
| KCNQ1 | CACNG4 | 0.667046 | 8 | 0 | 7 |
| KCNQ1 | CHRNB2 | 0.663711 | 6 | 0 | 5 |
| KCNQ1 | PIK3R5 | 0.662594 | 4 | 0 | 4 |
| KCNQ1 | CGA | 0.657232 | 4 | 0 | 4 |
| KCNQ1 | PNPLA2 | 0.654056 | 9 | 0 | 7 |
| KCNQ1 | OR2H2 | 0.653826 | 7 | 0 | 6 |
| KCNQ1 | FABP2 | 0.652424 | 4 | 0 | 3 |
| KCNQ1 | MAPT-AS1 | 0.651654 | 3 | 0 | 3 |
| KCNQ1 | PPP1R3F | 0.649079 | 4 | 0 | 3 |
| KCNQ1 | CDX2 | 0.648339 | 7 | 0 | 6 |
| KCNQ1 | CETN1 | 0.644351 | 5 | 0 | 5 |
| KCNQ1 | IL27 | 0.642235 | 3 | 0 | 3 |
| KCNQ1 | ATOH1 | 0.641686 | 6 | 0 | 4 |
| KCNQ1 | SLC4A1 | 0.641233 | 8 | 0 | 6 |
| KCNQ1 | PRG2 | 0.638469 | 6 | 0 | 5 |
| KCNQ1 | S1PR4 | 0.636646 | 5 | 0 | 4 |
| KCNQ1 | RAB3IL1 | 0.635374 | 8 | 0 | 6 |
| KCNQ1 | GJB4 | 0.634785 | 7 | 0 | 6 |
| KCNQ1 | CNTFR-AS1 | 0.633201 | 3 | 0 | 3 |
For details and further investigation, click here