| Full name: keratin 2 | Alias Symbol: KRTE | ||
| Type: protein-coding gene | Cytoband: 12q13.13 | ||
| Entrez ID: 3849 | HGNC ID: HGNC:6439 | Ensembl Gene: ENSG00000172867 | OMIM ID: 600194 |
| Drug and gene relationship at DGIdb | |||
Expression of KRT2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KRT2 | 3849 | 207908_at | -0.2366 | 0.4990 | |
| GSE20347 | KRT2 | 3849 | 207908_at | -0.2570 | 0.0139 | |
| GSE23400 | KRT2 | 3849 | 207908_at | -0.3927 | 0.0000 | |
| GSE26886 | KRT2 | 3849 | 207908_at | -1.5617 | 0.0001 | |
| GSE29001 | KRT2 | 3849 | 207908_at | -0.4971 | 0.0775 | |
| GSE38129 | KRT2 | 3849 | 207908_at | -0.4112 | 0.0043 | |
| GSE45670 | KRT2 | 3849 | 207908_at | -0.2913 | 0.4142 | |
| GSE53622 | KRT2 | 3849 | 10698 | -1.4443 | 0.0000 | |
| GSE53624 | KRT2 | 3849 | 10698 | -2.6278 | 0.0000 | |
| GSE63941 | KRT2 | 3849 | 207908_at | 0.0346 | 0.8754 | |
| GSE77861 | KRT2 | 3849 | 207908_at | -0.6896 | 0.0250 | |
| TCGA | KRT2 | 3849 | RNAseq | 3.8800 | 0.0012 |
Upregulated datasets: 1; Downregulated datasets: 3.
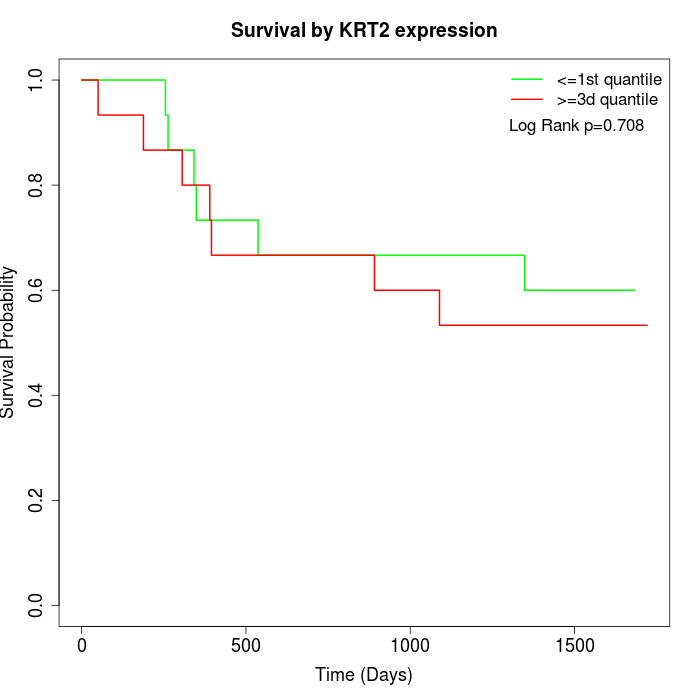
Survival by KRT2 expression:
Note: Click image to view full size file.
Copy number change of KRT2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KRT2 | 3849 | 8 | 1 | 21 | |
| GSE20123 | KRT2 | 3849 | 8 | 1 | 21 | |
| GSE43470 | KRT2 | 3849 | 3 | 0 | 40 | |
| GSE46452 | KRT2 | 3849 | 7 | 1 | 51 | |
| GSE47630 | KRT2 | 3849 | 10 | 2 | 28 | |
| GSE54993 | KRT2 | 3849 | 0 | 5 | 65 | |
| GSE54994 | KRT2 | 3849 | 5 | 1 | 47 | |
| GSE60625 | KRT2 | 3849 | 0 | 0 | 11 | |
| GSE74703 | KRT2 | 3849 | 3 | 0 | 33 | |
| GSE74704 | KRT2 | 3849 | 5 | 1 | 14 | |
| TCGA | KRT2 | 3849 | 13 | 11 | 72 |
Total number of gains: 62; Total number of losses: 23; Total Number of normals: 403.
Somatic mutations of KRT2:
Generating mutation plots.
Highly correlated genes for KRT2:
Showing top 20/1243 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KRT2 | ACER1 | 0.725432 | 4 | 0 | 4 |
| KRT2 | PAX8 | 0.722895 | 7 | 0 | 7 |
| KRT2 | LINC00423 | 0.717917 | 3 | 0 | 3 |
| KRT2 | RUNX1T1 | 0.715246 | 3 | 0 | 3 |
| KRT2 | SPEF2 | 0.714151 | 3 | 0 | 3 |
| KRT2 | IFNA21 | 0.710338 | 7 | 0 | 7 |
| KRT2 | PIWIL2 | 0.701976 | 6 | 0 | 6 |
| KRT2 | EPB42 | 0.696301 | 6 | 0 | 6 |
| KRT2 | MC5R | 0.692327 | 5 | 0 | 5 |
| KRT2 | FRAS1 | 0.682852 | 5 | 0 | 5 |
| KRT2 | ZNF460 | 0.675444 | 5 | 0 | 5 |
| KRT2 | COLEC10 | 0.674705 | 4 | 0 | 4 |
| KRT2 | CCR9 | 0.67453 | 4 | 0 | 4 |
| KRT2 | NOS1 | 0.673541 | 4 | 0 | 3 |
| KRT2 | PTPRT | 0.670893 | 4 | 0 | 3 |
| KRT2 | KRT78 | 0.667438 | 4 | 0 | 4 |
| KRT2 | PCBP1-AS1 | 0.666128 | 3 | 0 | 3 |
| KRT2 | EDA2R | 0.665777 | 6 | 0 | 5 |
| KRT2 | GNRHR | 0.664614 | 5 | 0 | 5 |
| KRT2 | SLC22A13 | 0.664611 | 4 | 0 | 4 |
For details and further investigation, click here