| Full name: kyphoscoliosis peptidase | Alias Symbol: FLJ33207 | ||
| Type: protein-coding gene | Cytoband: 3q22.2 | ||
| Entrez ID: 339855 | HGNC ID: HGNC:26576 | Ensembl Gene: ENSG00000174611 | OMIM ID: 605739 |
| Drug and gene relationship at DGIdb | |||
Expression of KY:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KY | 339855 | 1564254_at | 0.0924 | 0.6815 | |
| GSE26886 | KY | 339855 | 1564254_at | 0.1929 | 0.1084 | |
| GSE45670 | KY | 339855 | 1569991_at | 0.1027 | 0.2788 | |
| GSE53622 | KY | 339855 | 81929 | -0.0997 | 0.5105 | |
| GSE53624 | KY | 339855 | 81929 | -0.1233 | 0.3015 | |
| GSE63941 | KY | 339855 | 1564254_at | 0.0072 | 0.9703 | |
| GSE77861 | KY | 339855 | 1564254_at | 0.0498 | 0.7024 | |
| GSE97050 | KY | 339855 | A_23_P399897 | -0.7839 | 0.3291 | |
| SRP133303 | KY | 339855 | RNAseq | 0.1313 | 0.7147 | |
| SRP159526 | KY | 339855 | RNAseq | 0.1264 | 0.8155 | |
| SRP193095 | KY | 339855 | RNAseq | 1.6616 | 0.0000 | |
| TCGA | KY | 339855 | RNAseq | -0.9538 | 0.0496 |
Upregulated datasets: 1; Downregulated datasets: 0.
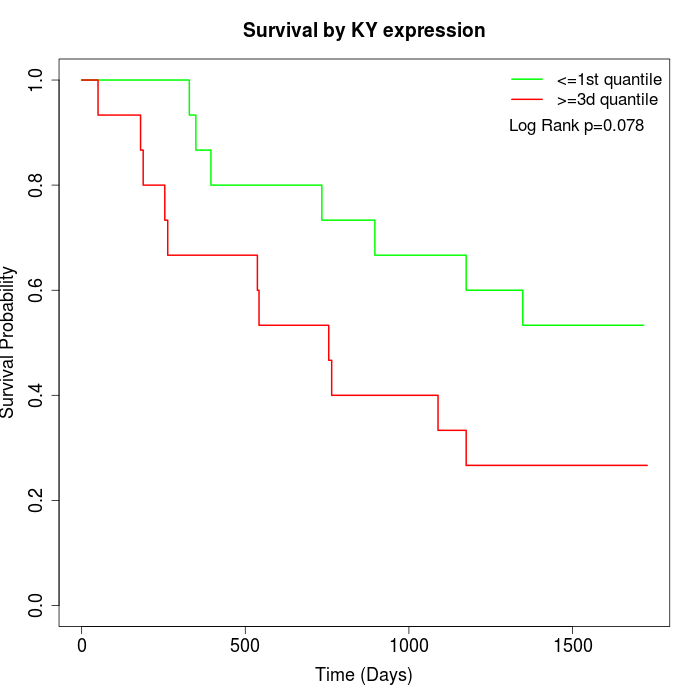
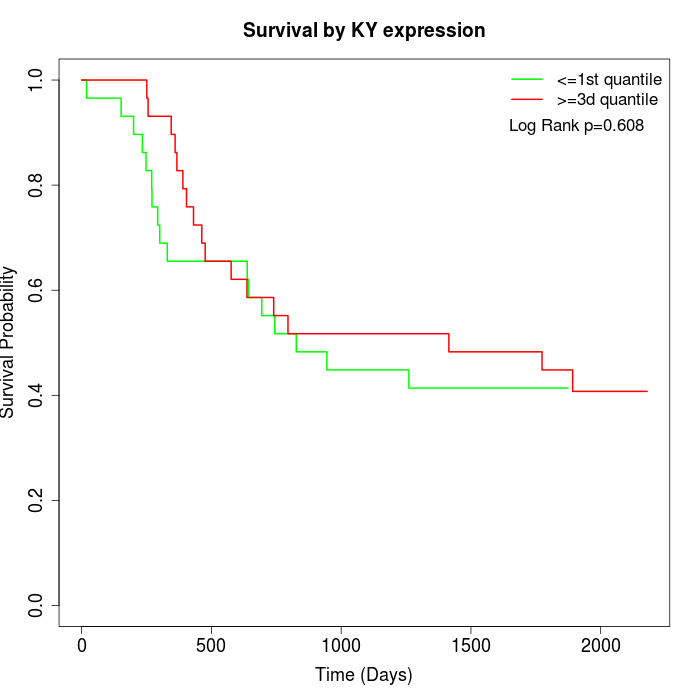
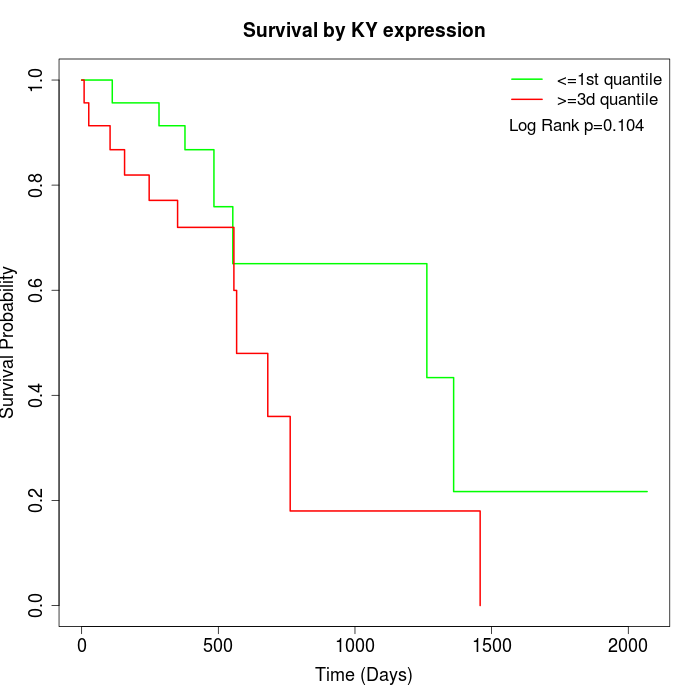
Survival by KY expression:
Note: Click image to view full size file.
Copy number change of KY:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KY | 339855 | 16 | 0 | 14 | |
| GSE20123 | KY | 339855 | 16 | 0 | 14 | |
| GSE43470 | KY | 339855 | 21 | 0 | 22 | |
| GSE46452 | KY | 339855 | 15 | 4 | 40 | |
| GSE47630 | KY | 339855 | 18 | 3 | 19 | |
| GSE54993 | KY | 339855 | 2 | 8 | 60 | |
| GSE54994 | KY | 339855 | 32 | 1 | 20 | |
| GSE60625 | KY | 339855 | 0 | 6 | 5 | |
| GSE74703 | KY | 339855 | 18 | 0 | 18 | |
| GSE74704 | KY | 339855 | 11 | 0 | 9 | |
| TCGA | KY | 339855 | 59 | 5 | 32 |
Total number of gains: 208; Total number of losses: 27; Total Number of normals: 253.
Somatic mutations of KY:
Generating mutation plots.
Highly correlated genes for KY:
Showing top 20/38 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KY | GSTM2 | 0.667017 | 3 | 0 | 3 |
| KY | ZNF483 | 0.632425 | 3 | 0 | 3 |
| KY | GSTM1 | 0.62778 | 3 | 0 | 3 |
| KY | FMR1NB | 0.61769 | 3 | 0 | 3 |
| KY | INPP5J | 0.617401 | 3 | 0 | 3 |
| KY | EXTL1 | 0.607283 | 3 | 0 | 3 |
| KY | WFIKKN1 | 0.592405 | 4 | 0 | 4 |
| KY | CRYGB | 0.584527 | 4 | 0 | 3 |
| KY | RIMS4 | 0.577848 | 4 | 0 | 3 |
| KY | GRK1 | 0.569288 | 3 | 0 | 3 |
| KY | SLC1A6 | 0.563582 | 4 | 0 | 3 |
| KY | MAP1LC3B2 | 0.560144 | 4 | 0 | 4 |
| KY | CRYBA1 | 0.558481 | 3 | 0 | 3 |
| KY | LRRC71 | 0.557034 | 3 | 0 | 3 |
| KY | CCL16 | 0.555514 | 4 | 0 | 3 |
| KY | HMX1 | 0.553469 | 5 | 0 | 3 |
| KY | UPK3A | 0.553248 | 5 | 0 | 3 |
| KY | DLGAP3 | 0.547134 | 4 | 0 | 3 |
| KY | OR7A17 | 0.542738 | 4 | 0 | 3 |
| KY | DCAF15 | 0.541714 | 4 | 0 | 3 |
For details and further investigation, click here