| Full name: phosphatidylinositol glycan anchor biosynthesis class B | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 15q21.3 | ||
| Entrez ID: 9488 | HGNC ID: HGNC:8959 | Ensembl Gene: ENSG00000069943 | OMIM ID: 604122 |
| Drug and gene relationship at DGIdb | |||
Expression of PIGB:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PIGB | 9488 | 205452_at | -0.4512 | 0.1977 | |
| GSE20347 | PIGB | 9488 | 205452_at | -0.3091 | 0.0505 | |
| GSE23400 | PIGB | 9488 | 205452_at | -0.0545 | 0.2869 | |
| GSE26886 | PIGB | 9488 | 205452_at | -0.3725 | 0.1322 | |
| GSE29001 | PIGB | 9488 | 205452_at | -0.3444 | 0.0991 | |
| GSE38129 | PIGB | 9488 | 205452_at | -0.2386 | 0.0409 | |
| GSE45670 | PIGB | 9488 | 205452_at | -0.1349 | 0.3778 | |
| GSE53622 | PIGB | 9488 | 78025 | -0.1783 | 0.0346 | |
| GSE53624 | PIGB | 9488 | 78025 | -0.3009 | 0.0148 | |
| GSE63941 | PIGB | 9488 | 205452_at | -0.4685 | 0.1613 | |
| GSE77861 | PIGB | 9488 | 205452_at | -0.3750 | 0.2061 | |
| GSE97050 | PIGB | 9488 | A_23_P77174 | 0.0082 | 0.9731 | |
| SRP007169 | PIGB | 9488 | RNAseq | -0.4295 | 0.4603 | |
| SRP008496 | PIGB | 9488 | RNAseq | -0.3367 | 0.3504 | |
| SRP064894 | PIGB | 9488 | RNAseq | -0.1082 | 0.3888 | |
| SRP133303 | PIGB | 9488 | RNAseq | -0.1828 | 0.1893 | |
| SRP159526 | PIGB | 9488 | RNAseq | -0.7944 | 0.0398 | |
| SRP193095 | PIGB | 9488 | RNAseq | -0.3483 | 0.0001 | |
| SRP219564 | PIGB | 9488 | RNAseq | 0.0042 | 0.9866 | |
| TCGA | PIGB | 9488 | RNAseq | 0.0291 | 0.6444 |
Upregulated datasets: 0; Downregulated datasets: 0.
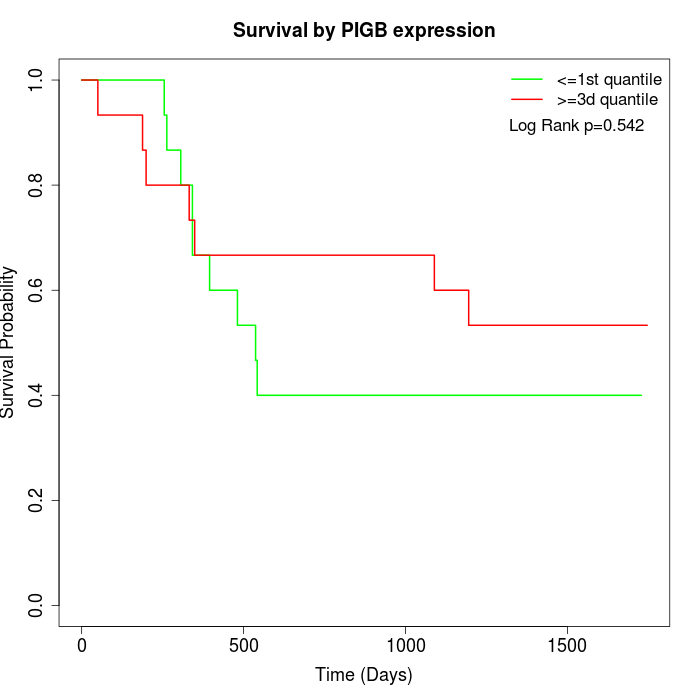
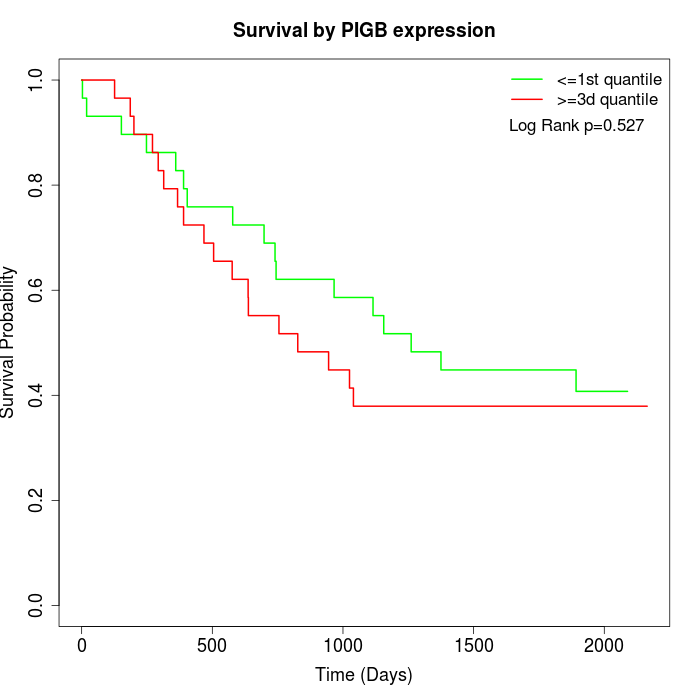
Survival by PIGB expression:
Note: Click image to view full size file.
Copy number change of PIGB:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PIGB | 9488 | 6 | 3 | 21 | |
| GSE20123 | PIGB | 9488 | 6 | 3 | 21 | |
| GSE43470 | PIGB | 9488 | 4 | 3 | 36 | |
| GSE46452 | PIGB | 9488 | 3 | 7 | 49 | |
| GSE47630 | PIGB | 9488 | 8 | 10 | 22 | |
| GSE54993 | PIGB | 9488 | 5 | 6 | 59 | |
| GSE54994 | PIGB | 9488 | 5 | 8 | 40 | |
| GSE60625 | PIGB | 9488 | 4 | 0 | 7 | |
| GSE74703 | PIGB | 9488 | 4 | 2 | 30 | |
| GSE74704 | PIGB | 9488 | 2 | 3 | 15 | |
| TCGA | PIGB | 9488 | 10 | 16 | 70 |
Total number of gains: 57; Total number of losses: 61; Total Number of normals: 370.
Somatic mutations of PIGB:
Generating mutation plots.
Highly correlated genes for PIGB:
Showing top 20/590 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PIGB | ULK3 | 0.765527 | 3 | 0 | 3 |
| PIGB | ZNF398 | 0.755937 | 3 | 0 | 3 |
| PIGB | DHDDS | 0.746785 | 3 | 0 | 3 |
| PIGB | ZC3H13 | 0.726417 | 3 | 0 | 3 |
| PIGB | UBQLN1 | 0.725548 | 3 | 0 | 3 |
| PIGB | NPAT | 0.724615 | 4 | 0 | 4 |
| PIGB | FAR1 | 0.718312 | 3 | 0 | 3 |
| PIGB | KLF4 | 0.717019 | 3 | 0 | 3 |
| PIGB | COG5 | 0.713794 | 3 | 0 | 3 |
| PIGB | FAM98C | 0.71116 | 3 | 0 | 3 |
| PIGB | YPEL3 | 0.70878 | 3 | 0 | 3 |
| PIGB | BAG4 | 0.705671 | 3 | 0 | 3 |
| PIGB | LATS1 | 0.703065 | 3 | 0 | 3 |
| PIGB | RAD17 | 0.702666 | 5 | 0 | 5 |
| PIGB | PSD3 | 0.700838 | 3 | 0 | 3 |
| PIGB | PRADC1 | 0.696818 | 3 | 0 | 3 |
| PIGB | DPH3 | 0.695925 | 3 | 0 | 3 |
| PIGB | FGD4 | 0.69391 | 4 | 0 | 3 |
| PIGB | PANK1 | 0.693467 | 5 | 0 | 5 |
| PIGB | CHIC2 | 0.692915 | 4 | 0 | 3 |
For details and further investigation, click here