| Full name: LCK proto-oncogene, Src family tyrosine kinase | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 1p35.2 | ||
| Entrez ID: 3932 | HGNC ID: HGNC:6524 | Ensembl Gene: ENSG00000182866 | OMIM ID: 153390 |
| Drug and gene relationship at DGIdb | |||
LCK involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04380 | Osteoclast differentiation | |
| hsa04650 | Natural killer cell mediated cytotoxicity | |
| hsa04660 | T cell receptor signaling pathway |
Expression of LCK:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LCK | 3932 | 204891_s_at | 0.6933 | 0.5859 | |
| GSE20347 | LCK | 3932 | 204891_s_at | 0.0327 | 0.9078 | |
| GSE23400 | LCK | 3932 | 204891_s_at | 0.2017 | 0.0524 | |
| GSE26886 | LCK | 3932 | 204891_s_at | 0.2106 | 0.6507 | |
| GSE29001 | LCK | 3932 | 204891_s_at | 0.0484 | 0.8686 | |
| GSE38129 | LCK | 3932 | 204891_s_at | 0.1059 | 0.7250 | |
| GSE45670 | LCK | 3932 | 204891_s_at | 0.2141 | 0.5027 | |
| GSE53622 | LCK | 3932 | 98541 | 0.2320 | 0.1271 | |
| GSE53624 | LCK | 3932 | 98541 | -0.0912 | 0.6116 | |
| GSE63941 | LCK | 3932 | 204890_s_at | 0.0025 | 0.9934 | |
| GSE77861 | LCK | 3932 | 204891_s_at | -0.0869 | 0.6561 | |
| GSE97050 | LCK | 3932 | A_33_P3382746 | 1.0590 | 0.0996 | |
| SRP007169 | LCK | 3932 | RNAseq | -0.6221 | 0.4219 | |
| SRP064894 | LCK | 3932 | RNAseq | 0.2638 | 0.3580 | |
| SRP133303 | LCK | 3932 | RNAseq | -0.3007 | 0.4181 | |
| SRP159526 | LCK | 3932 | RNAseq | -0.9674 | 0.2297 | |
| SRP193095 | LCK | 3932 | RNAseq | -0.1555 | 0.5483 | |
| SRP219564 | LCK | 3932 | RNAseq | 0.2117 | 0.7016 | |
| TCGA | LCK | 3932 | RNAseq | 0.3225 | 0.1029 |
Upregulated datasets: 0; Downregulated datasets: 0.
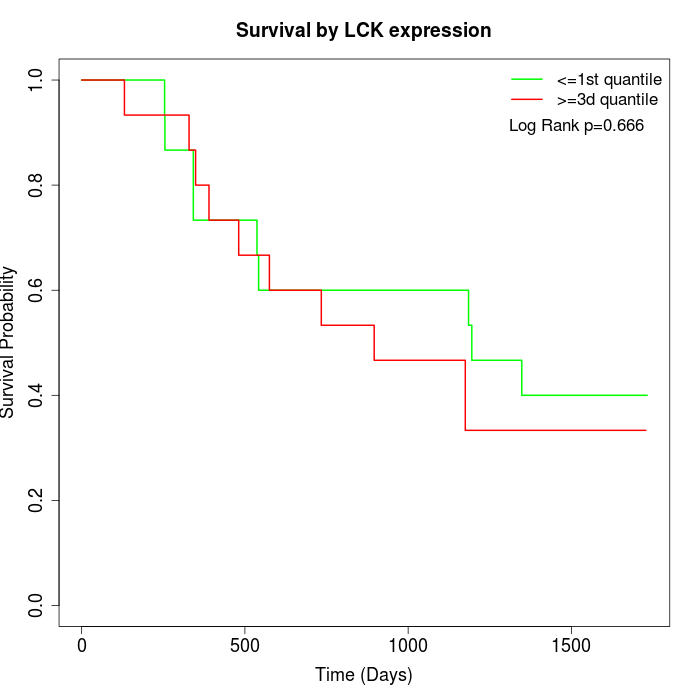
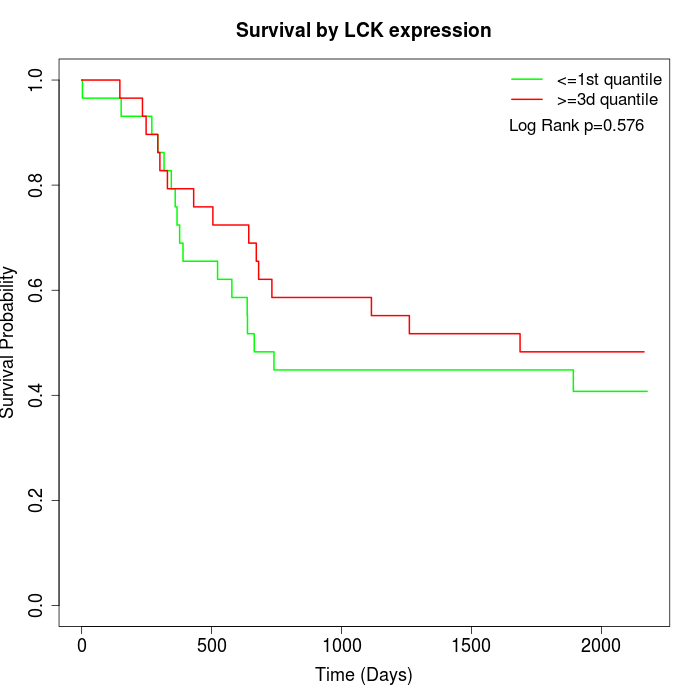
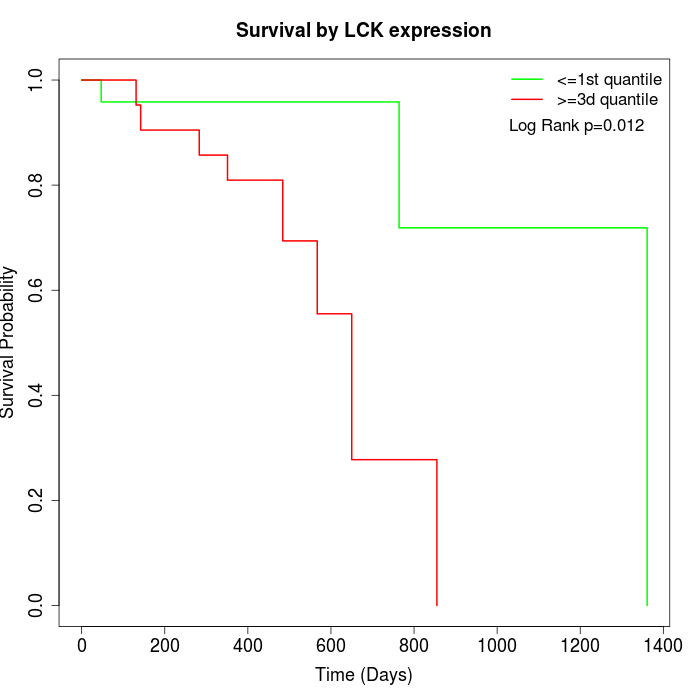
Survival by LCK expression:
Note: Click image to view full size file.
Copy number change of LCK:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LCK | 3932 | 1 | 5 | 24 | |
| GSE20123 | LCK | 3932 | 1 | 4 | 25 | |
| GSE43470 | LCK | 3932 | 6 | 4 | 33 | |
| GSE46452 | LCK | 3932 | 5 | 1 | 53 | |
| GSE47630 | LCK | 3932 | 8 | 3 | 29 | |
| GSE54993 | LCK | 3932 | 1 | 1 | 68 | |
| GSE54994 | LCK | 3932 | 11 | 2 | 40 | |
| GSE60625 | LCK | 3932 | 0 | 0 | 11 | |
| GSE74703 | LCK | 3932 | 4 | 3 | 29 | |
| GSE74704 | LCK | 3932 | 0 | 0 | 20 | |
| TCGA | LCK | 3932 | 10 | 20 | 66 |
Total number of gains: 47; Total number of losses: 43; Total Number of normals: 398.
Somatic mutations of LCK:
Generating mutation plots.
Highly correlated genes for LCK:
Showing top 20/420 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LCK | CCR4 | 0.783315 | 3 | 0 | 3 |
| LCK | IL12RB1 | 0.782269 | 5 | 0 | 5 |
| LCK | CD5 | 0.772688 | 4 | 0 | 4 |
| LCK | CD3D | 0.761946 | 12 | 0 | 11 |
| LCK | P2RY8 | 0.753715 | 4 | 0 | 4 |
| LCK | APBB1IP | 0.74906 | 6 | 0 | 6 |
| LCK | SLA | 0.749033 | 10 | 0 | 10 |
| LCK | CORO1A | 0.746586 | 12 | 0 | 10 |
| LCK | SASH3 | 0.743851 | 11 | 0 | 10 |
| LCK | TNFAIP8L2 | 0.742091 | 6 | 0 | 6 |
| LCK | IL2RB | 0.739691 | 11 | 0 | 10 |
| LCK | C16orf54 | 0.739443 | 6 | 0 | 6 |
| LCK | POU2AF1 | 0.735735 | 3 | 0 | 3 |
| LCK | SNX20 | 0.71725 | 7 | 0 | 6 |
| LCK | CALML5 | 0.714887 | 3 | 0 | 3 |
| LCK | CYTIP | 0.712334 | 11 | 0 | 9 |
| LCK | LAPTM5 | 0.71112 | 10 | 0 | 9 |
| LCK | FCGR2A | 0.706858 | 3 | 0 | 3 |
| LCK | CD2 | 0.704231 | 12 | 0 | 11 |
| LCK | ARHGAP9 | 0.70367 | 7 | 0 | 6 |
For details and further investigation, click here