| Full name: long intergenic non-protein coding RNA 574 | Alias Symbol: FLJ13162|dJ182D15.1|CRALA | ||
| Type: non-coding RNA | Cytoband: 6q27 | ||
| Entrez ID: 80069 | HGNC ID: HGNC:21598 | Ensembl Gene: ENSG00000231690 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of LINC00574:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LINC00574 | 80069 | 220904_at | 0.0097 | 0.9774 | |
| GSE20347 | LINC00574 | 80069 | 220904_at | -0.0092 | 0.9391 | |
| GSE23400 | LINC00574 | 80069 | 220904_at | -0.1554 | 0.0035 | |
| GSE26886 | LINC00574 | 80069 | 220904_at | -0.0658 | 0.7244 | |
| GSE29001 | LINC00574 | 80069 | 220904_at | 0.0370 | 0.8742 | |
| GSE38129 | LINC00574 | 80069 | 220904_at | -0.0812 | 0.3147 | |
| GSE45670 | LINC00574 | 80069 | 220904_at | 0.1300 | 0.2915 | |
| GSE53622 | LINC00574 | 80069 | 117482 | -0.0481 | 0.6386 | |
| GSE53624 | LINC00574 | 80069 | 26503 | 0.0627 | 0.4244 | |
| GSE63941 | LINC00574 | 80069 | 220904_at | -0.3277 | 0.0723 | |
| GSE77861 | LINC00574 | 80069 | 220904_at | 0.0718 | 0.6351 |
Upregulated datasets: 0; Downregulated datasets: 0.
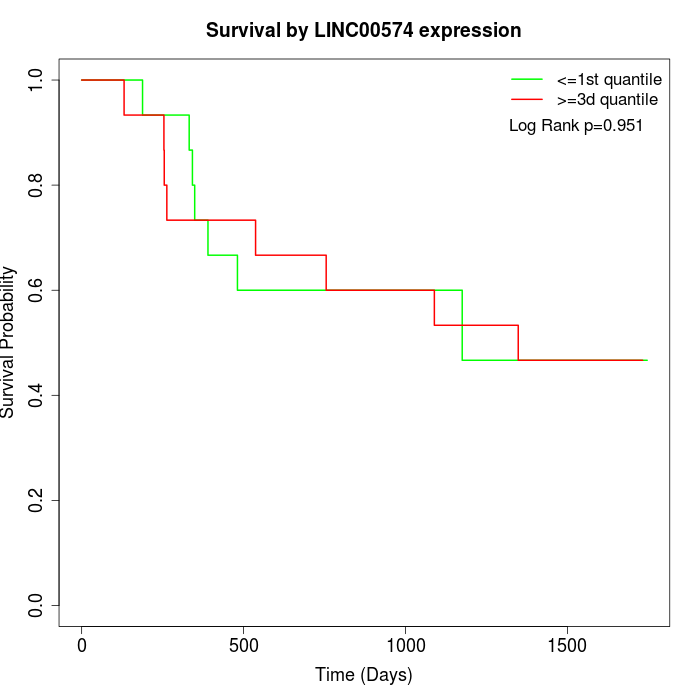
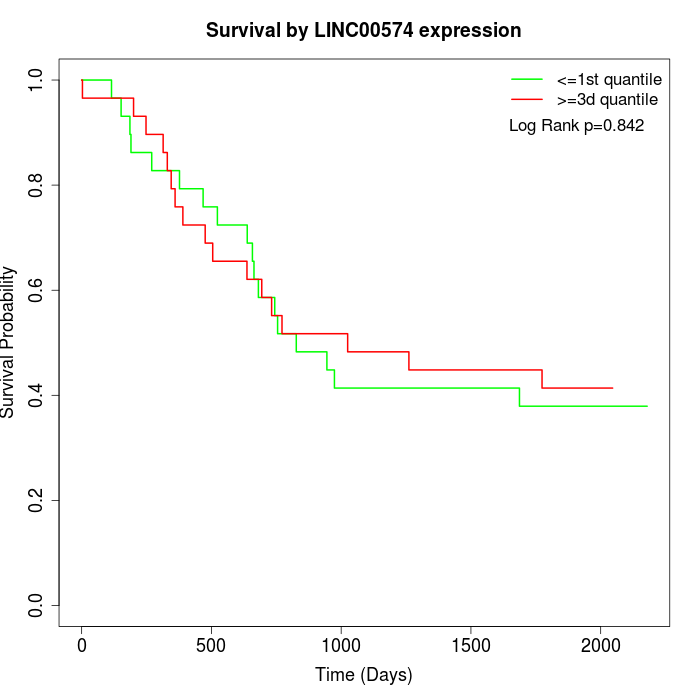
Survival by LINC00574 expression:
Note: Click image to view full size file.
Copy number change of LINC00574:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LINC00574 | 80069 | 4 | 6 | 20 | |
| GSE20123 | LINC00574 | 80069 | 4 | 6 | 20 | |
| GSE43470 | LINC00574 | 80069 | 2 | 0 | 41 | |
| GSE46452 | LINC00574 | 80069 | 3 | 10 | 46 | |
| GSE47630 | LINC00574 | 80069 | 7 | 7 | 26 | |
| GSE54993 | LINC00574 | 80069 | 4 | 3 | 63 | |
| GSE54994 | LINC00574 | 80069 | 10 | 8 | 35 | |
| GSE60625 | LINC00574 | 80069 | 1 | 1 | 9 | |
| GSE74703 | LINC00574 | 80069 | 2 | 0 | 34 | |
| GSE74704 | LINC00574 | 80069 | 2 | 3 | 15 | |
| TCGA | LINC00574 | 80069 | 14 | 20 | 62 |
Total number of gains: 53; Total number of losses: 64; Total Number of normals: 371.
Somatic mutations of LINC00574:
Generating mutation plots.
Highly correlated genes for LINC00574:
Showing top 20/614 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LINC00574 | NOVA2 | 0.715397 | 4 | 0 | 3 |
| LINC00574 | KIR2DL4 | 0.693593 | 5 | 0 | 5 |
| LINC00574 | GAD2 | 0.687482 | 5 | 0 | 5 |
| LINC00574 | CRYGC | 0.674339 | 3 | 0 | 3 |
| LINC00574 | C1orf194 | 0.662269 | 3 | 0 | 3 |
| LINC00574 | PROKR2 | 0.65952 | 3 | 0 | 3 |
| LINC00574 | NEU3 | 0.658811 | 4 | 0 | 4 |
| LINC00574 | CXCR1 | 0.656482 | 7 | 0 | 7 |
| LINC00574 | KCNA1 | 0.656052 | 5 | 0 | 5 |
| LINC00574 | TACR3 | 0.653743 | 5 | 0 | 5 |
| LINC00574 | SV2C | 0.650011 | 4 | 0 | 4 |
| LINC00574 | CYP19A1 | 0.647811 | 4 | 0 | 3 |
| LINC00574 | KCNH6 | 0.645164 | 4 | 0 | 3 |
| LINC00574 | USP2 | 0.644742 | 4 | 0 | 4 |
| LINC00574 | CNR2 | 0.641032 | 5 | 0 | 5 |
| LINC00574 | NTNG1 | 0.638217 | 4 | 0 | 4 |
| LINC00574 | HECW1 | 0.638016 | 5 | 0 | 5 |
| LINC00574 | ATG10 | 0.637976 | 4 | 0 | 4 |
| LINC00574 | MMP16 | 0.637876 | 3 | 0 | 3 |
| LINC00574 | NRGN | 0.637806 | 5 | 0 | 4 |
For details and further investigation, click here