| Full name: prokineticin receptor 2 | Alias Symbol: GPR73b|PKR2|GPRg2|dJ680N4.3 | ||
| Type: protein-coding gene | Cytoband: 20p12.3 | ||
| Entrez ID: 128674 | HGNC ID: HGNC:15836 | Ensembl Gene: ENSG00000101292 | OMIM ID: 607123 |
| Drug and gene relationship at DGIdb | |||
Expression of PROKR2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PROKR2 | 128674 | 234450_at | -0.1222 | 0.7324 | |
| GSE26886 | PROKR2 | 128674 | 234450_at | 0.0416 | 0.7698 | |
| GSE45670 | PROKR2 | 128674 | 234450_at | 0.1450 | 0.1781 | |
| GSE53622 | PROKR2 | 128674 | 64669 | -0.3398 | 0.0002 | |
| GSE53624 | PROKR2 | 128674 | 64669 | -0.4451 | 0.0000 | |
| GSE63941 | PROKR2 | 128674 | 234450_at | 0.0959 | 0.5862 | |
| GSE77861 | PROKR2 | 128674 | 234450_at | -0.0206 | 0.8215 | |
| SRP133303 | PROKR2 | 128674 | RNAseq | 0.2963 | 0.1260 | |
| SRP193095 | PROKR2 | 128674 | RNAseq | 0.2064 | 0.1897 | |
| TCGA | PROKR2 | 128674 | RNAseq | -1.0575 | 0.5566 |
Upregulated datasets: 0; Downregulated datasets: 0.
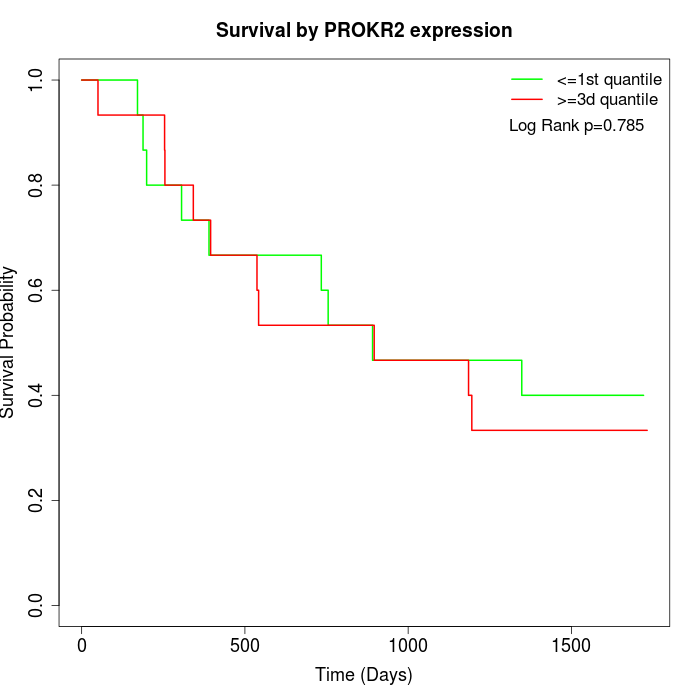
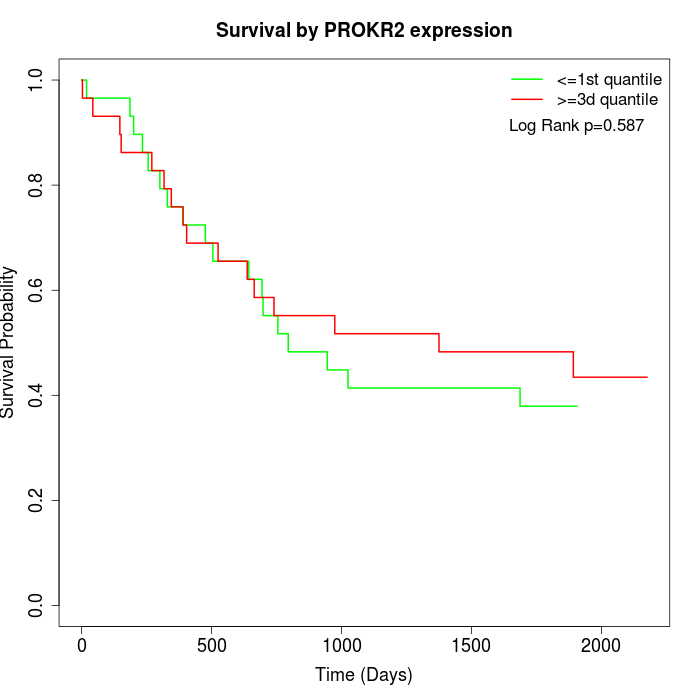
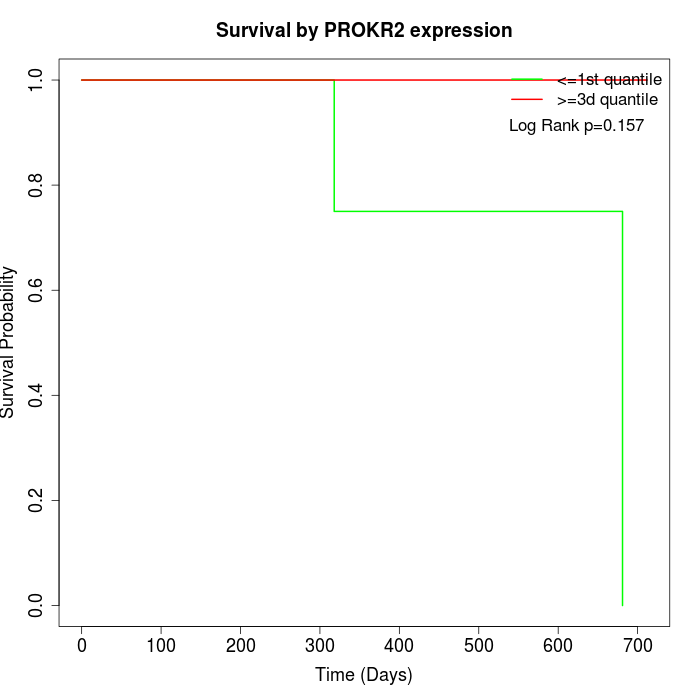
Survival by PROKR2 expression:
Note: Click image to view full size file.
Copy number change of PROKR2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PROKR2 | 128674 | 10 | 3 | 17 | |
| GSE20123 | PROKR2 | 128674 | 10 | 3 | 17 | |
| GSE43470 | PROKR2 | 128674 | 10 | 1 | 32 | |
| GSE46452 | PROKR2 | 128674 | 26 | 1 | 32 | |
| GSE47630 | PROKR2 | 128674 | 18 | 4 | 18 | |
| GSE54993 | PROKR2 | 128674 | 2 | 14 | 54 | |
| GSE54994 | PROKR2 | 128674 | 25 | 2 | 26 | |
| GSE60625 | PROKR2 | 128674 | 0 | 0 | 11 | |
| GSE74703 | PROKR2 | 128674 | 9 | 1 | 26 | |
| GSE74704 | PROKR2 | 128674 | 5 | 1 | 14 | |
| TCGA | PROKR2 | 128674 | 40 | 11 | 45 |
Total number of gains: 155; Total number of losses: 41; Total Number of normals: 292.
Somatic mutations of PROKR2:
Generating mutation plots.
Highly correlated genes for PROKR2:
Showing top 20/38 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PROKR2 | LINC00574 | 0.65952 | 3 | 0 | 3 |
| PROKR2 | KCNJ13 | 0.65742 | 3 | 0 | 3 |
| PROKR2 | ZNF491 | 0.651152 | 3 | 0 | 3 |
| PROKR2 | GFRA2 | 0.640503 | 3 | 0 | 3 |
| PROKR2 | ADAM7 | 0.618472 | 4 | 0 | 3 |
| PROKR2 | OR4D2 | 0.606546 | 5 | 0 | 5 |
| PROKR2 | C22orf42 | 0.605088 | 3 | 0 | 3 |
| PROKR2 | SLC13A2 | 0.6027 | 3 | 0 | 3 |
| PROKR2 | MUC8 | 0.597361 | 3 | 0 | 3 |
| PROKR2 | CA4 | 0.595877 | 4 | 0 | 3 |
| PROKR2 | HRG | 0.593637 | 3 | 0 | 3 |
| PROKR2 | TEC | 0.587451 | 3 | 0 | 3 |
| PROKR2 | TSSK3 | 0.586085 | 5 | 0 | 4 |
| PROKR2 | OR2J3 | 0.573385 | 3 | 0 | 3 |
| PROKR2 | CRYGC | 0.568712 | 5 | 0 | 4 |
| PROKR2 | RAB4B | 0.568179 | 4 | 0 | 3 |
| PROKR2 | C20orf173 | 0.567428 | 3 | 0 | 3 |
| PROKR2 | EPHA6 | 0.564512 | 4 | 0 | 3 |
| PROKR2 | PCA3 | 0.560586 | 4 | 0 | 3 |
| PROKR2 | CR1 | 0.556437 | 3 | 0 | 3 |
For details and further investigation, click here