| Full name: lemur tyrosine kinase 2 | Alias Symbol: KIAA1079|KPI2|KPI-2|cprk|LMR2|BREK|AATYK2|PPP1R100 | ||
| Type: protein-coding gene | Cytoband: 7q21.3 | ||
| Entrez ID: 22853 | HGNC ID: HGNC:17880 | Ensembl Gene: ENSG00000164715 | OMIM ID: 610989 |
| Drug and gene relationship at DGIdb | |||
Expression of LMTK2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LMTK2 | 22853 | 226375_at | 0.2414 | 0.7265 | |
| GSE20347 | LMTK2 | 22853 | 206223_at | -0.1086 | 0.2008 | |
| GSE23400 | LMTK2 | 22853 | 206223_at | -0.2059 | 0.0000 | |
| GSE26886 | LMTK2 | 22853 | 226375_at | -0.5685 | 0.0115 | |
| GSE29001 | LMTK2 | 22853 | 206223_at | -0.0879 | 0.6299 | |
| GSE38129 | LMTK2 | 22853 | 206223_at | -0.1188 | 0.1011 | |
| GSE45670 | LMTK2 | 22853 | 226375_at | 0.1633 | 0.3996 | |
| GSE53622 | LMTK2 | 22853 | 43065 | -0.0441 | 0.7096 | |
| GSE53624 | LMTK2 | 22853 | 43065 | -0.4409 | 0.0000 | |
| GSE63941 | LMTK2 | 22853 | 226375_at | 0.3769 | 0.3988 | |
| GSE77861 | LMTK2 | 22853 | 226375_at | 0.3519 | 0.0716 | |
| GSE97050 | LMTK2 | 22853 | A_33_P3266923 | -0.0072 | 0.9812 | |
| SRP007169 | LMTK2 | 22853 | RNAseq | -0.4615 | 0.3448 | |
| SRP008496 | LMTK2 | 22853 | RNAseq | -0.5023 | 0.0540 | |
| SRP064894 | LMTK2 | 22853 | RNAseq | -0.9939 | 0.0010 | |
| SRP133303 | LMTK2 | 22853 | RNAseq | 0.0408 | 0.8389 | |
| SRP159526 | LMTK2 | 22853 | RNAseq | -0.4642 | 0.1888 | |
| SRP193095 | LMTK2 | 22853 | RNAseq | 0.0088 | 0.9594 | |
| SRP219564 | LMTK2 | 22853 | RNAseq | -0.3622 | 0.4891 | |
| TCGA | LMTK2 | 22853 | RNAseq | -0.0183 | 0.7950 |
Upregulated datasets: 0; Downregulated datasets: 0.
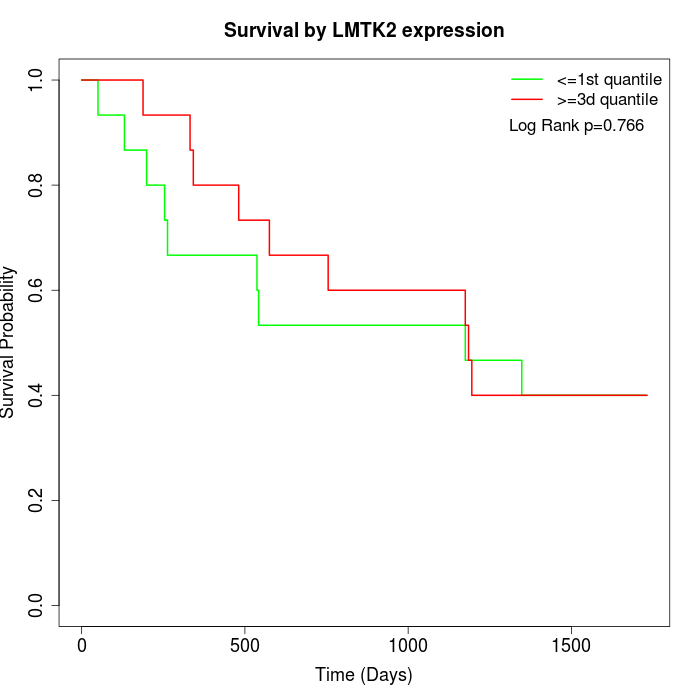
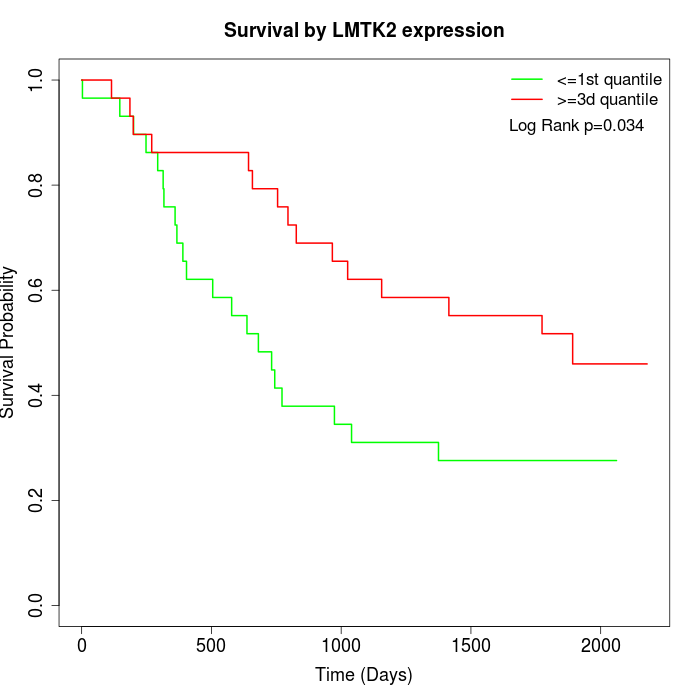
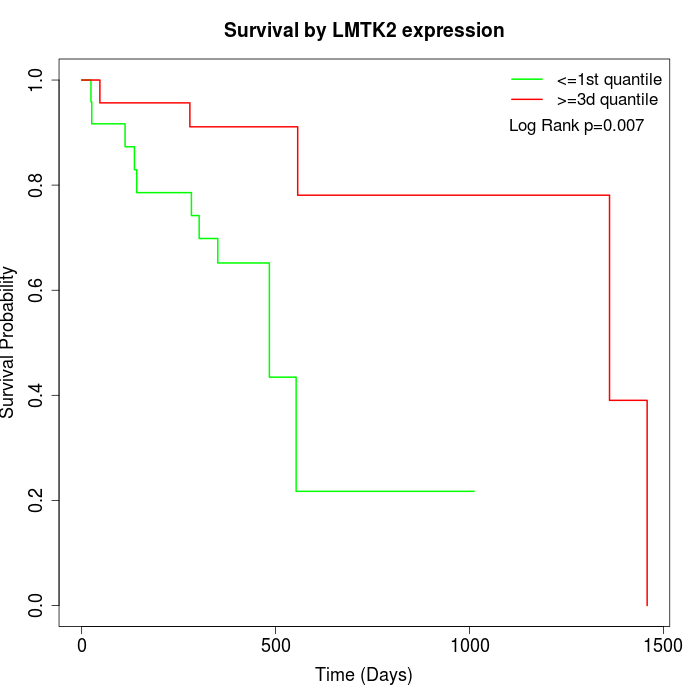
Survival by LMTK2 expression:
Note: Click image to view full size file.
Copy number change of LMTK2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LMTK2 | 22853 | 12 | 0 | 18 | |
| GSE20123 | LMTK2 | 22853 | 12 | 0 | 18 | |
| GSE43470 | LMTK2 | 22853 | 6 | 2 | 35 | |
| GSE46452 | LMTK2 | 22853 | 12 | 0 | 47 | |
| GSE47630 | LMTK2 | 22853 | 9 | 2 | 29 | |
| GSE54993 | LMTK2 | 22853 | 1 | 9 | 60 | |
| GSE54994 | LMTK2 | 22853 | 16 | 3 | 34 | |
| GSE60625 | LMTK2 | 22853 | 0 | 0 | 11 | |
| GSE74703 | LMTK2 | 22853 | 6 | 1 | 29 | |
| GSE74704 | LMTK2 | 22853 | 9 | 0 | 11 | |
| TCGA | LMTK2 | 22853 | 55 | 4 | 37 |
Total number of gains: 138; Total number of losses: 21; Total Number of normals: 329.
Somatic mutations of LMTK2:
Generating mutation plots.
Highly correlated genes for LMTK2:
Showing top 20/163 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LMTK2 | KDM7A | 0.738234 | 3 | 0 | 3 |
| LMTK2 | BROX | 0.708721 | 3 | 0 | 3 |
| LMTK2 | TET2 | 0.670587 | 4 | 0 | 3 |
| LMTK2 | TRDN | 0.6637 | 3 | 0 | 3 |
| LMTK2 | MRPL37 | 0.662548 | 3 | 0 | 3 |
| LMTK2 | STK25 | 0.661738 | 3 | 0 | 3 |
| LMTK2 | PRLR | 0.655253 | 3 | 0 | 3 |
| LMTK2 | CAPNS2 | 0.645334 | 3 | 0 | 3 |
| LMTK2 | ZNF117 | 0.64288 | 3 | 0 | 3 |
| LMTK2 | LAMC3 | 0.63813 | 4 | 0 | 3 |
| LMTK2 | ARMC10 | 0.635661 | 3 | 0 | 3 |
| LMTK2 | DLGAP2 | 0.63068 | 5 | 0 | 5 |
| LMTK2 | MED8 | 0.629103 | 3 | 0 | 3 |
| LMTK2 | RLIM | 0.625633 | 3 | 0 | 3 |
| LMTK2 | SLC22A23 | 0.617159 | 5 | 0 | 3 |
| LMTK2 | MAP4K5 | 0.616698 | 3 | 0 | 3 |
| LMTK2 | BAIAP2L1 | 0.616482 | 8 | 0 | 6 |
| LMTK2 | SMURF1 | 0.615982 | 7 | 0 | 6 |
| LMTK2 | AAMP | 0.613885 | 4 | 0 | 3 |
| LMTK2 | ZNF343 | 0.613485 | 3 | 0 | 3 |
For details and further investigation, click here