| Full name: lysophosphatidylcholine acyltransferase 2 | Alias Symbol: FLJ20481|AGPAT11|LysoPAFAT | ||
| Type: protein-coding gene | Cytoband: 16q12.2 | ||
| Entrez ID: 54947 | HGNC ID: HGNC:26032 | Ensembl Gene: ENSG00000087253 | OMIM ID: 612040 |
| Drug and gene relationship at DGIdb | |||
Expression of LPCAT2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LPCAT2 | 54947 | 227889_at | -0.1641 | 0.8708 | |
| GSE26886 | LPCAT2 | 54947 | 227889_at | 0.0903 | 0.8411 | |
| GSE45670 | LPCAT2 | 54947 | 227889_at | 0.2583 | 0.4130 | |
| GSE53622 | LPCAT2 | 54947 | 34033 | 0.4473 | 0.0000 | |
| GSE53624 | LPCAT2 | 54947 | 34033 | 0.3873 | 0.0002 | |
| GSE63941 | LPCAT2 | 54947 | 239598_s_at | 2.0230 | 0.0011 | |
| GSE77861 | LPCAT2 | 54947 | 227889_at | 0.4060 | 0.4695 | |
| GSE97050 | LPCAT2 | 54947 | A_23_P141021 | 0.7354 | 0.0809 | |
| SRP007169 | LPCAT2 | 54947 | RNAseq | 0.0680 | 0.8259 | |
| SRP008496 | LPCAT2 | 54947 | RNAseq | 0.5553 | 0.0955 | |
| SRP064894 | LPCAT2 | 54947 | RNAseq | 0.5335 | 0.0217 | |
| SRP133303 | LPCAT2 | 54947 | RNAseq | 0.5727 | 0.0016 | |
| SRP159526 | LPCAT2 | 54947 | RNAseq | 0.1341 | 0.7005 | |
| SRP193095 | LPCAT2 | 54947 | RNAseq | 0.1435 | 0.3221 | |
| SRP219564 | LPCAT2 | 54947 | RNAseq | 0.9486 | 0.1150 | |
| TCGA | LPCAT2 | 54947 | RNAseq | 0.2131 | 0.0103 |
Upregulated datasets: 1; Downregulated datasets: 0.
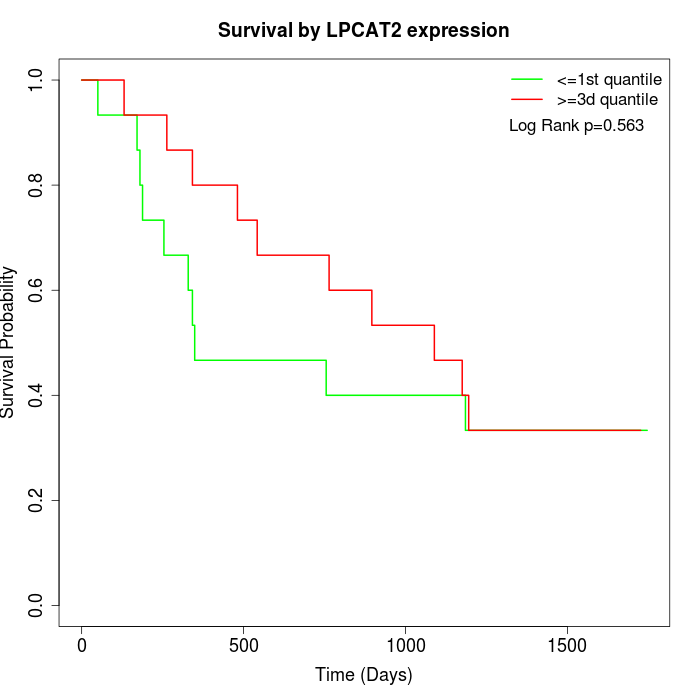
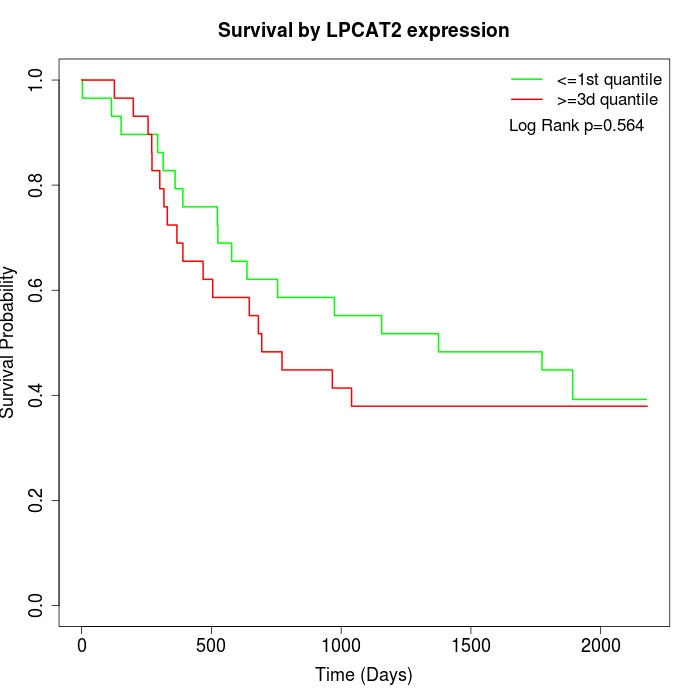
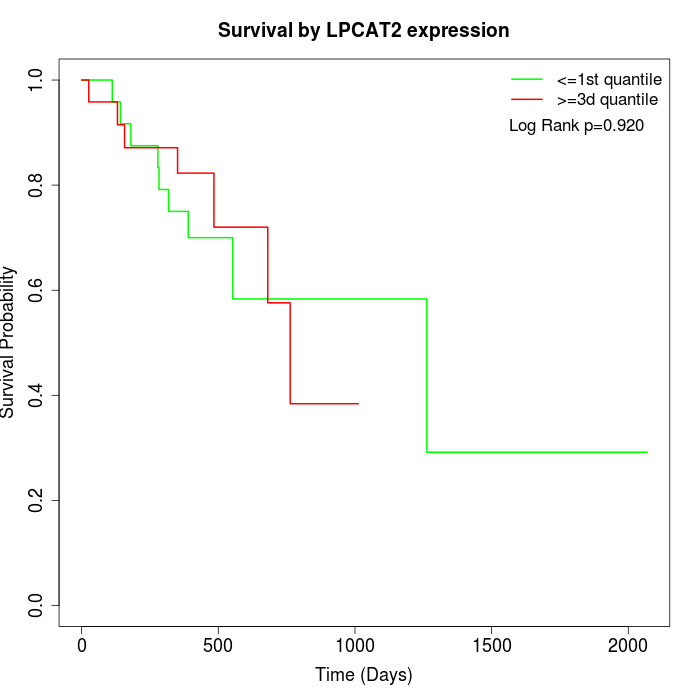
Survival by LPCAT2 expression:
Note: Click image to view full size file.
Copy number change of LPCAT2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LPCAT2 | 54947 | 4 | 0 | 26 | |
| GSE20123 | LPCAT2 | 54947 | 4 | 0 | 26 | |
| GSE43470 | LPCAT2 | 54947 | 2 | 7 | 34 | |
| GSE46452 | LPCAT2 | 54947 | 38 | 1 | 20 | |
| GSE47630 | LPCAT2 | 54947 | 10 | 8 | 22 | |
| GSE54993 | LPCAT2 | 54947 | 2 | 4 | 64 | |
| GSE54994 | LPCAT2 | 54947 | 7 | 10 | 36 | |
| GSE60625 | LPCAT2 | 54947 | 4 | 0 | 7 | |
| GSE74703 | LPCAT2 | 54947 | 2 | 4 | 30 | |
| GSE74704 | LPCAT2 | 54947 | 3 | 0 | 17 | |
| TCGA | LPCAT2 | 54947 | 25 | 12 | 59 |
Total number of gains: 101; Total number of losses: 46; Total Number of normals: 341.
Somatic mutations of LPCAT2:
Generating mutation plots.
Highly correlated genes for LPCAT2:
Showing top 20/211 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LPCAT2 | RHBDF2 | 0.768262 | 3 | 0 | 3 |
| LPCAT2 | FANCL | 0.753294 | 3 | 0 | 3 |
| LPCAT2 | FLRT2 | 0.751556 | 3 | 0 | 3 |
| LPCAT2 | MYOF | 0.748867 | 3 | 0 | 3 |
| LPCAT2 | NCAPG | 0.73309 | 3 | 0 | 3 |
| LPCAT2 | MRPL46 | 0.729887 | 3 | 0 | 3 |
| LPCAT2 | TUBGCP3 | 0.728429 | 3 | 0 | 3 |
| LPCAT2 | PPP6R3 | 0.728029 | 3 | 0 | 3 |
| LPCAT2 | SP4 | 0.726057 | 3 | 0 | 3 |
| LPCAT2 | GPSM2 | 0.722474 | 3 | 0 | 3 |
| LPCAT2 | NCAPD3 | 0.720474 | 3 | 0 | 3 |
| LPCAT2 | LRRC8A | 0.71999 | 3 | 0 | 3 |
| LPCAT2 | LRRCC1 | 0.719397 | 3 | 0 | 3 |
| LPCAT2 | ZCCHC8 | 0.719107 | 4 | 0 | 4 |
| LPCAT2 | RELA | 0.714095 | 3 | 0 | 3 |
| LPCAT2 | MFN1 | 0.70717 | 3 | 0 | 3 |
| LPCAT2 | DEPDC4 | 0.706924 | 3 | 0 | 3 |
| LPCAT2 | RAC1 | 0.705967 | 3 | 0 | 3 |
| LPCAT2 | RNF144B | 0.702428 | 3 | 0 | 3 |
| LPCAT2 | NDUFV1 | 0.701435 | 3 | 0 | 3 |
For details and further investigation, click here