| Full name: fibronectin leucine rich transmembrane protein 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 14q31.3 | ||
| Entrez ID: 23768 | HGNC ID: HGNC:3761 | Ensembl Gene: ENSG00000185070 | OMIM ID: 604807 |
| Drug and gene relationship at DGIdb | |||
Expression of FLRT2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | FLRT2 | 23768 | 112756 | 0.7678 | 0.0000 | |
| GSE53624 | FLRT2 | 23768 | 112756 | 0.7038 | 0.0002 | |
| GSE97050 | FLRT2 | 23768 | A_24_P236235 | 1.0556 | 0.0555 | |
| SRP007169 | FLRT2 | 23768 | RNAseq | 3.8499 | 0.0000 | |
| SRP008496 | FLRT2 | 23768 | RNAseq | 3.6195 | 0.0000 | |
| SRP064894 | FLRT2 | 23768 | RNAseq | 1.8260 | 0.0000 | |
| SRP133303 | FLRT2 | 23768 | RNAseq | 1.1383 | 0.0043 | |
| SRP159526 | FLRT2 | 23768 | RNAseq | 1.1621 | 0.0622 | |
| SRP193095 | FLRT2 | 23768 | RNAseq | 2.1264 | 0.0000 | |
| SRP219564 | FLRT2 | 23768 | RNAseq | 1.7738 | 0.0145 | |
| TCGA | FLRT2 | 23768 | RNAseq | 0.3029 | 0.0900 |
Upregulated datasets: 6; Downregulated datasets: 0.
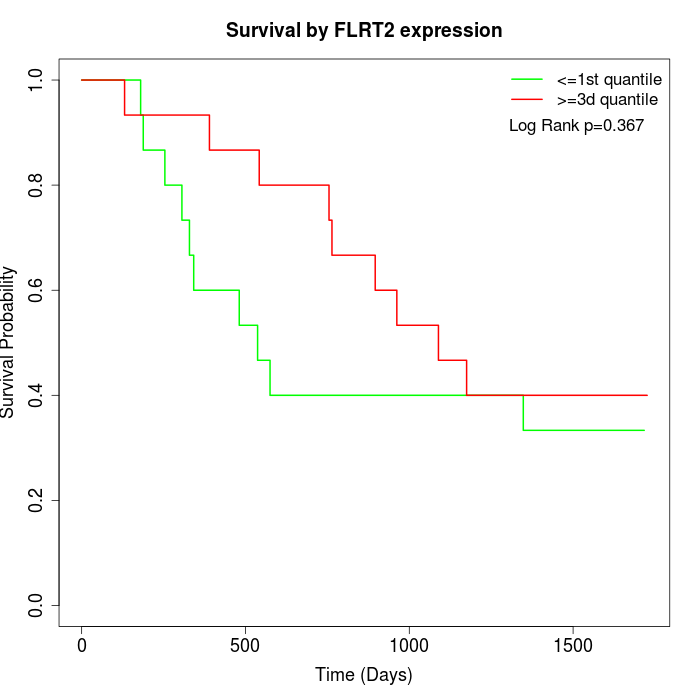
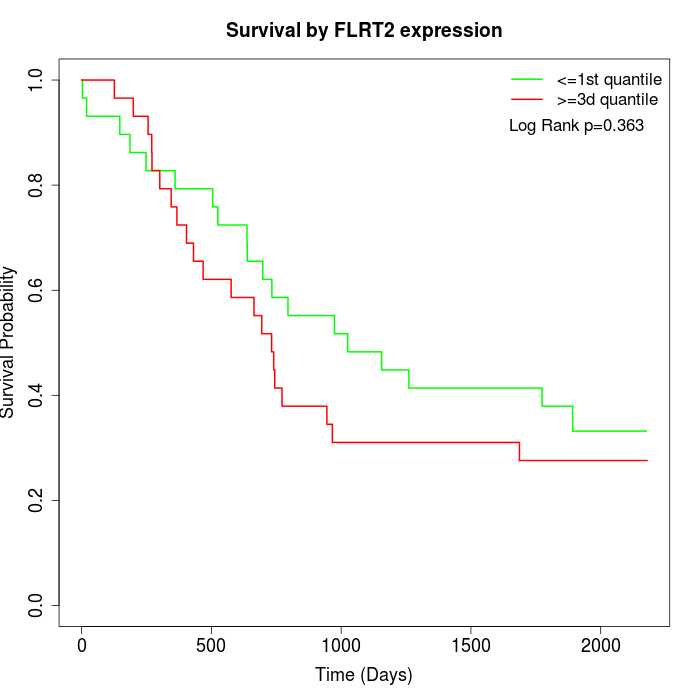
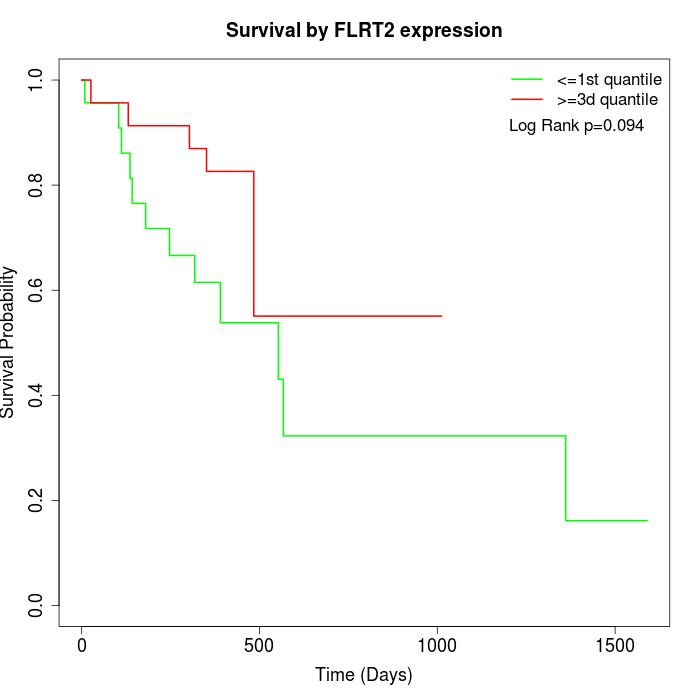
Survival by FLRT2 expression:
Note: Click image to view full size file.
Copy number change of FLRT2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FLRT2 | 23768 | 7 | 4 | 19 | |
| GSE20123 | FLRT2 | 23768 | 7 | 4 | 19 | |
| GSE43470 | FLRT2 | 23768 | 8 | 3 | 32 | |
| GSE46452 | FLRT2 | 23768 | 16 | 3 | 40 | |
| GSE47630 | FLRT2 | 23768 | 11 | 8 | 21 | |
| GSE54993 | FLRT2 | 23768 | 3 | 8 | 59 | |
| GSE54994 | FLRT2 | 23768 | 19 | 4 | 30 | |
| GSE60625 | FLRT2 | 23768 | 0 | 2 | 9 | |
| GSE74703 | FLRT2 | 23768 | 6 | 3 | 27 | |
| GSE74704 | FLRT2 | 23768 | 4 | 4 | 12 | |
| TCGA | FLRT2 | 23768 | 31 | 21 | 44 |
Total number of gains: 112; Total number of losses: 64; Total Number of normals: 312.
Somatic mutations of FLRT2:
Generating mutation plots.
Highly correlated genes for FLRT2:
Showing top 20/32 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FLRT2 | LPCAT2 | 0.751556 | 3 | 0 | 3 |
| FLRT2 | THSD1 | 0.72007 | 3 | 0 | 3 |
| FLRT2 | TANC2 | 0.715422 | 3 | 0 | 3 |
| FLRT2 | AMIGO2 | 0.714512 | 3 | 0 | 3 |
| FLRT2 | KLF7 | 0.694848 | 3 | 0 | 3 |
| FLRT2 | GUCY1A2 | 0.684002 | 3 | 0 | 3 |
| FLRT2 | JKAMP | 0.68028 | 3 | 0 | 3 |
| FLRT2 | GLT8D2 | 0.677748 | 3 | 0 | 3 |
| FLRT2 | PROCR | 0.657195 | 3 | 0 | 3 |
| FLRT2 | COL5A3 | 0.654911 | 3 | 0 | 3 |
| FLRT2 | MMP16 | 0.648436 | 3 | 0 | 3 |
| FLRT2 | PRRX1 | 0.647981 | 3 | 0 | 3 |
| FLRT2 | TDO2 | 0.647649 | 3 | 0 | 3 |
| FLRT2 | SEC23A | 0.642816 | 3 | 0 | 3 |
| FLRT2 | AFAP1L1 | 0.640177 | 3 | 0 | 3 |
| FLRT2 | VEGFC | 0.633413 | 4 | 0 | 4 |
| FLRT2 | CAV3 | 0.625166 | 3 | 0 | 3 |
| FLRT2 | GPX8 | 0.624927 | 3 | 0 | 3 |
| FLRT2 | DCBLD1 | 0.622216 | 3 | 0 | 3 |
| FLRT2 | CYP27C1 | 0.616857 | 4 | 0 | 3 |
For details and further investigation, click here