| Full name: RELA proto-oncogene, NF-kB subunit | Alias Symbol: p65 | ||
| Type: protein-coding gene | Cytoband: 11q13.1 | ||
| Entrez ID: 5970 | HGNC ID: HGNC:9955 | Ensembl Gene: ENSG00000173039 | OMIM ID: 164014 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
RELA involved pathways:
Expression of RELA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RELA | 5970 | 201783_s_at | -0.0540 | 0.8865 | |
| GSE20347 | RELA | 5970 | 201783_s_at | 0.0165 | 0.9231 | |
| GSE23400 | RELA | 5970 | 201783_s_at | -0.0564 | 0.2515 | |
| GSE26886 | RELA | 5970 | 201783_s_at | 0.1896 | 0.3599 | |
| GSE29001 | RELA | 5970 | 201783_s_at | -0.3778 | 0.0234 | |
| GSE38129 | RELA | 5970 | 201783_s_at | -0.1036 | 0.3411 | |
| GSE45670 | RELA | 5970 | 201783_s_at | -0.1640 | 0.2066 | |
| GSE53622 | RELA | 5970 | 52082 | -0.1009 | 0.2008 | |
| GSE53624 | RELA | 5970 | 67171 | 0.1011 | 0.1001 | |
| GSE63941 | RELA | 5970 | 201783_s_at | 0.2312 | 0.5098 | |
| GSE77861 | RELA | 5970 | 201783_s_at | 0.3183 | 0.0618 | |
| GSE97050 | RELA | 5970 | A_33_P3209433 | 0.1104 | 0.6173 | |
| SRP007169 | RELA | 5970 | RNAseq | 0.1110 | 0.8215 | |
| SRP008496 | RELA | 5970 | RNAseq | -0.0338 | 0.9066 | |
| SRP064894 | RELA | 5970 | RNAseq | -0.2769 | 0.0758 | |
| SRP133303 | RELA | 5970 | RNAseq | 0.0942 | 0.6655 | |
| SRP159526 | RELA | 5970 | RNAseq | -0.3604 | 0.0355 | |
| SRP193095 | RELA | 5970 | RNAseq | 0.1105 | 0.3048 | |
| SRP219564 | RELA | 5970 | RNAseq | -0.3622 | 0.1890 | |
| TCGA | RELA | 5970 | RNAseq | 0.0286 | 0.5055 |
Upregulated datasets: 0; Downregulated datasets: 0.
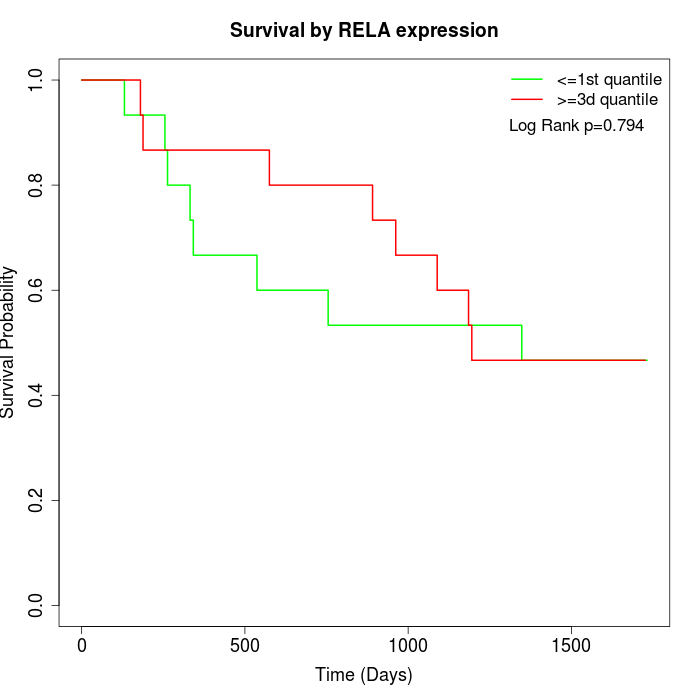
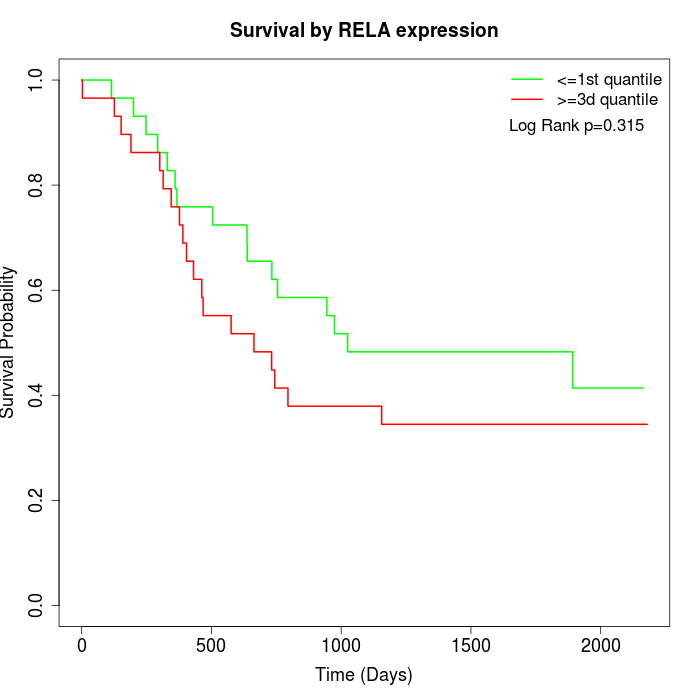
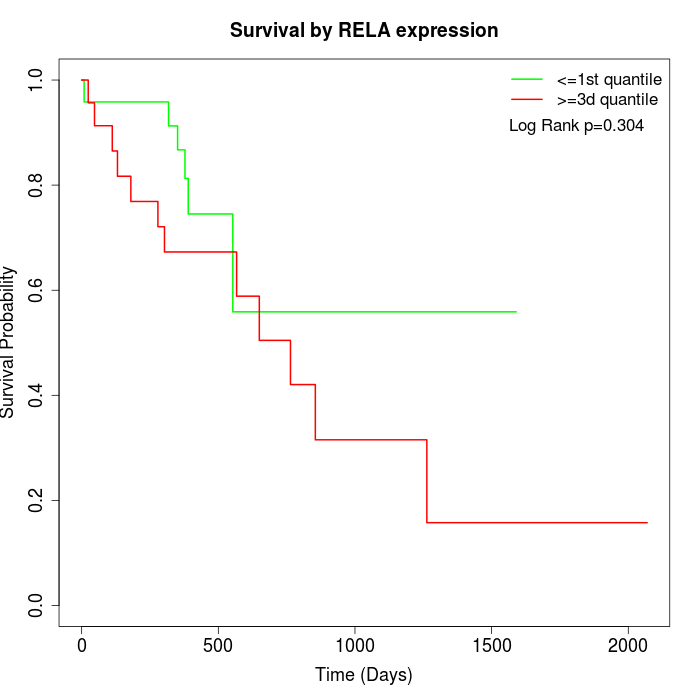
Survival by RELA expression:
Note: Click image to view full size file.
Copy number change of RELA:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RELA | 5970 | 8 | 5 | 17 | |
| GSE20123 | RELA | 5970 | 8 | 5 | 17 | |
| GSE43470 | RELA | 5970 | 4 | 1 | 38 | |
| GSE46452 | RELA | 5970 | 11 | 3 | 45 | |
| GSE47630 | RELA | 5970 | 7 | 5 | 28 | |
| GSE54993 | RELA | 5970 | 3 | 0 | 67 | |
| GSE54994 | RELA | 5970 | 7 | 5 | 41 | |
| GSE60625 | RELA | 5970 | 0 | 3 | 8 | |
| GSE74703 | RELA | 5970 | 3 | 0 | 33 | |
| GSE74704 | RELA | 5970 | 6 | 3 | 11 | |
| TCGA | RELA | 5970 | 23 | 7 | 66 |
Total number of gains: 80; Total number of losses: 37; Total Number of normals: 371.
Somatic mutations of RELA:
Generating mutation plots.
Highly correlated genes for RELA:
Showing top 20/143 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RELA | CIC | 0.759192 | 3 | 0 | 3 |
| RELA | S100A6 | 0.744684 | 4 | 0 | 4 |
| RELA | CTNNBL1 | 0.73717 | 3 | 0 | 3 |
| RELA | ZDHHC9 | 0.729849 | 3 | 0 | 3 |
| RELA | LPCAT2 | 0.714095 | 3 | 0 | 3 |
| RELA | PORCN | 0.710382 | 3 | 0 | 3 |
| RELA | RNF25 | 0.708926 | 3 | 0 | 3 |
| RELA | FLNB | 0.703268 | 3 | 0 | 3 |
| RELA | SIK1 | 0.697642 | 3 | 0 | 3 |
| RELA | PLTP | 0.693018 | 3 | 0 | 3 |
| RELA | TNFRSF12A | 0.691234 | 3 | 0 | 3 |
| RELA | RGS14 | 0.690212 | 3 | 0 | 3 |
| RELA | C19orf54 | 0.685547 | 4 | 0 | 3 |
| RELA | PMF1 | 0.683204 | 3 | 0 | 3 |
| RELA | BMP2 | 0.681703 | 3 | 0 | 3 |
| RELA | PXN | 0.678754 | 4 | 0 | 3 |
| RELA | SPIRE1 | 0.678269 | 3 | 0 | 3 |
| RELA | MRPL55 | 0.675858 | 3 | 0 | 3 |
| RELA | FTSJ1 | 0.670031 | 3 | 0 | 3 |
| RELA | AQR | 0.665504 | 3 | 0 | 3 |
For details and further investigation, click here