| Full name: small lysine rich protein 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 7q32.1 | ||
| Entrez ID: 100287482 | HGNC ID: HGNC:43561 | Ensembl Gene: ENSG00000240204 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of SMKR1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SMKR1 | 100287482 | 235736_at | 0.9507 | 0.0671 | |
| GSE26886 | SMKR1 | 100287482 | 235736_at | 0.9931 | 0.0201 | |
| GSE45670 | SMKR1 | 100287482 | 235736_at | 1.2287 | 0.0154 | |
| GSE53622 | SMKR1 | 100287482 | 97345 | 0.9861 | 0.0000 | |
| GSE53624 | SMKR1 | 100287482 | 111538 | 1.4520 | 0.0000 | |
| GSE63941 | SMKR1 | 100287482 | 235736_at | 0.7118 | 0.3777 | |
| GSE77861 | SMKR1 | 100287482 | 235736_at | 0.3376 | 0.1214 | |
| SRP159526 | SMKR1 | 100287482 | RNAseq | 3.1049 | 0.0000 |
Upregulated datasets: 3; Downregulated datasets: 0.
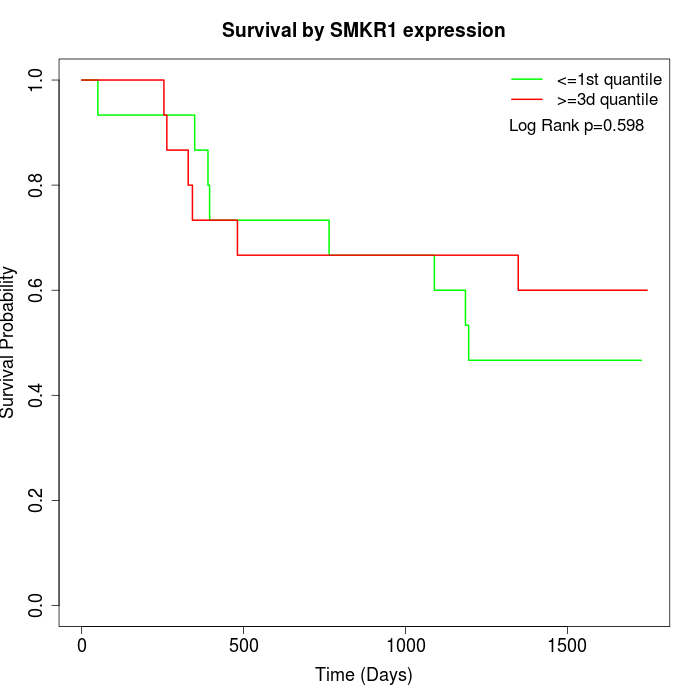
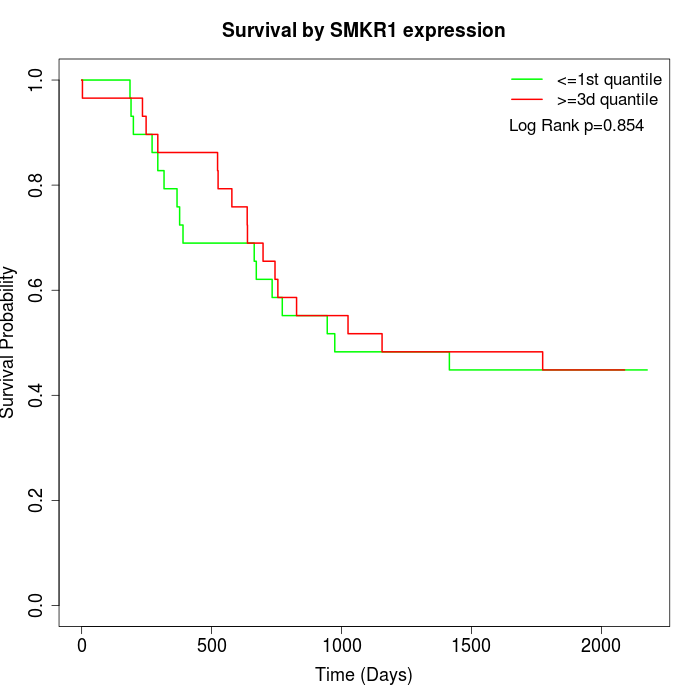
Survival by SMKR1 expression:
Note: Click image to view full size file.
Copy number change of SMKR1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SMKR1 | 100287482 | 8 | 1 | 21 | |
| GSE20123 | SMKR1 | 100287482 | 8 | 1 | 21 | |
| GSE43470 | SMKR1 | 100287482 | 2 | 3 | 38 | |
| GSE46452 | SMKR1 | 100287482 | 7 | 2 | 50 | |
| GSE47630 | SMKR1 | 100287482 | 7 | 4 | 29 | |
| GSE54993 | SMKR1 | 100287482 | 3 | 5 | 62 | |
| GSE54994 | SMKR1 | 100287482 | 8 | 7 | 38 | |
| GSE60625 | SMKR1 | 100287482 | 0 | 0 | 11 | |
| GSE74703 | SMKR1 | 100287482 | 2 | 3 | 31 | |
| GSE74704 | SMKR1 | 100287482 | 4 | 1 | 15 | |
| TCGA | SMKR1 | 100287482 | 29 | 21 | 46 |
Total number of gains: 78; Total number of losses: 48; Total Number of normals: 362.
Somatic mutations of SMKR1:
Generating mutation plots.
Highly correlated genes for SMKR1:
Showing top 20/630 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SMKR1 | DDX11 | 0.736114 | 3 | 0 | 3 |
| SMKR1 | LRRC58 | 0.718267 | 3 | 0 | 3 |
| SMKR1 | KHSRP | 0.717985 | 3 | 0 | 3 |
| SMKR1 | TNFAIP8L1 | 0.70932 | 3 | 0 | 3 |
| SMKR1 | TNPO2 | 0.704506 | 3 | 0 | 3 |
| SMKR1 | BLM | 0.700547 | 5 | 0 | 4 |
| SMKR1 | RBM28 | 0.696301 | 3 | 0 | 3 |
| SMKR1 | DESI2 | 0.689578 | 6 | 0 | 6 |
| SMKR1 | CHAF1B | 0.684225 | 3 | 0 | 3 |
| SMKR1 | RAD51AP1 | 0.682505 | 5 | 0 | 5 |
| SMKR1 | FLVCR1 | 0.681699 | 6 | 0 | 6 |
| SMKR1 | TMEM97 | 0.680538 | 5 | 0 | 4 |
| SMKR1 | PDCD2L | 0.680427 | 5 | 0 | 4 |
| SMKR1 | GPX4 | 0.680197 | 3 | 0 | 3 |
| SMKR1 | MRPS12 | 0.678134 | 4 | 0 | 3 |
| SMKR1 | KIAA0100 | 0.675288 | 4 | 0 | 3 |
| SMKR1 | RFX5 | 0.674754 | 3 | 0 | 3 |
| SMKR1 | STRADA | 0.673903 | 3 | 0 | 3 |
| SMKR1 | SPAG5 | 0.671664 | 5 | 0 | 4 |
| SMKR1 | ZNF384 | 0.670659 | 3 | 0 | 3 |
For details and further investigation, click here