| Full name: desmin | Alias Symbol: CMD1I|CSM1|CSM2 | ||
| Type: protein-coding gene | Cytoband: 2q35 | ||
| Entrez ID: 1674 | HGNC ID: HGNC:2770 | Ensembl Gene: ENSG00000175084 | OMIM ID: 125660 |
| Drug and gene relationship at DGIdb | |||
Expression of DES:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | DES | 1674 | 202222_s_at | -2.6431 | 0.2107 | |
| GSE20347 | DES | 1674 | 202222_s_at | -0.9492 | 0.0701 | |
| GSE23400 | DES | 1674 | 202222_s_at | -1.2138 | 0.0004 | |
| GSE26886 | DES | 1674 | 202222_s_at | 0.0767 | 0.7894 | |
| GSE29001 | DES | 1674 | 202222_s_at | -0.1734 | 0.5679 | |
| GSE38129 | DES | 1674 | 202222_s_at | -1.4271 | 0.0367 | |
| GSE45670 | DES | 1674 | 202222_s_at | -2.1071 | 0.0000 | |
| GSE53622 | DES | 1674 | 142791 | -3.3042 | 0.0000 | |
| GSE53624 | DES | 1674 | 142791 | -2.9941 | 0.0000 | |
| GSE63941 | DES | 1674 | 202222_s_at | -0.1798 | 0.3140 | |
| GSE77861 | DES | 1674 | 202222_s_at | 0.0646 | 0.7470 | |
| GSE97050 | DES | 1674 | A_33_P3275801 | -2.5162 | 0.0708 | |
| SRP064894 | DES | 1674 | RNAseq | -1.2858 | 0.0142 | |
| SRP133303 | DES | 1674 | RNAseq | -3.5670 | 0.0000 | |
| SRP159526 | DES | 1674 | RNAseq | -1.4215 | 0.2021 | |
| SRP219564 | DES | 1674 | RNAseq | -2.0231 | 0.1606 | |
| TCGA | DES | 1674 | RNAseq | -0.9206 | 0.0080 |
Upregulated datasets: 0; Downregulated datasets: 7.
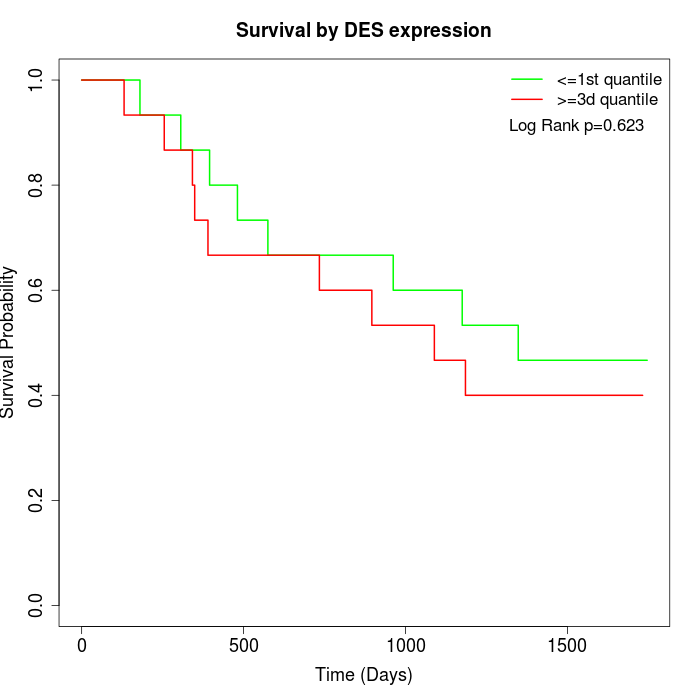
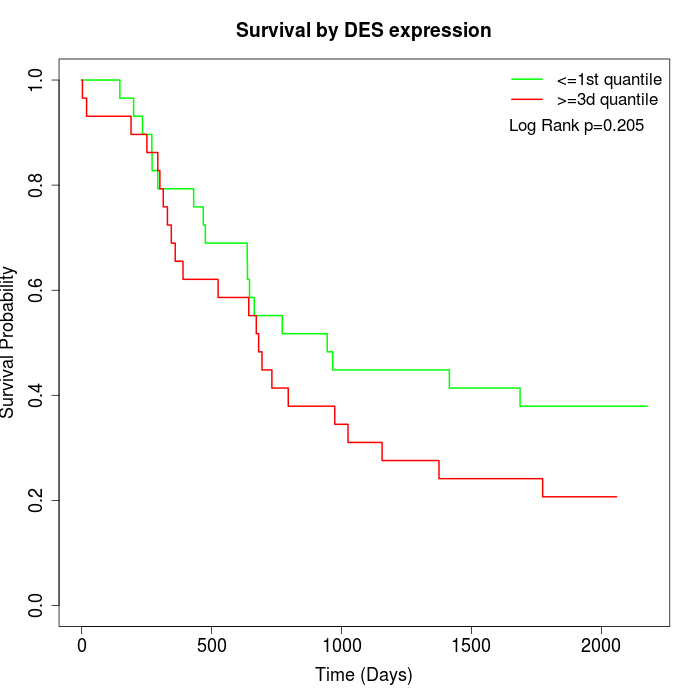
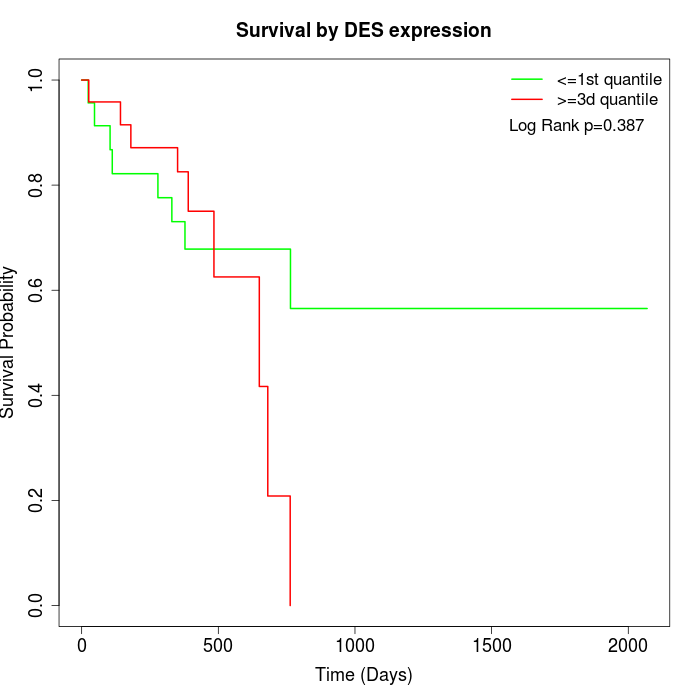
Survival by DES expression:
Note: Click image to view full size file.
Copy number change of DES:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | DES | 1674 | 2 | 14 | 14 | |
| GSE20123 | DES | 1674 | 2 | 13 | 15 | |
| GSE43470 | DES | 1674 | 1 | 6 | 36 | |
| GSE46452 | DES | 1674 | 0 | 5 | 54 | |
| GSE47630 | DES | 1674 | 4 | 5 | 31 | |
| GSE54993 | DES | 1674 | 1 | 2 | 67 | |
| GSE54994 | DES | 1674 | 8 | 9 | 36 | |
| GSE60625 | DES | 1674 | 0 | 3 | 8 | |
| GSE74703 | DES | 1674 | 1 | 5 | 30 | |
| GSE74704 | DES | 1674 | 2 | 7 | 11 | |
| TCGA | DES | 1674 | 13 | 25 | 58 |
Total number of gains: 34; Total number of losses: 94; Total Number of normals: 360.
Somatic mutations of DES:
Generating mutation plots.
Highly correlated genes for DES:
Showing top 20/1057 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| DES | LMOD1 | 0.858333 | 13 | 0 | 12 |
| DES | HMHB1 | 0.851969 | 3 | 0 | 3 |
| DES | CNN1 | 0.844739 | 11 | 0 | 10 |
| DES | PPP1R12B | 0.840345 | 11 | 0 | 10 |
| DES | ANGPTL1 | 0.838474 | 6 | 0 | 6 |
| DES | MYL9 | 0.828713 | 11 | 0 | 11 |
| DES | MYH11 | 0.828302 | 12 | 0 | 12 |
| DES | DAAM2 | 0.823302 | 9 | 0 | 9 |
| DES | ACTG2 | 0.820681 | 13 | 0 | 12 |
| DES | ASB5 | 0.817016 | 5 | 0 | 5 |
| DES | SYNM | 0.806782 | 12 | 0 | 11 |
| DES | ACTBL2 | 0.803398 | 3 | 0 | 3 |
| DES | AOC3 | 0.798535 | 12 | 0 | 12 |
| DES | PLN | 0.797172 | 13 | 0 | 12 |
| DES | PCP4 | 0.796772 | 11 | 0 | 11 |
| DES | ADCY5 | 0.793397 | 5 | 0 | 5 |
| DES | PRUNE2 | 0.791734 | 13 | 0 | 12 |
| DES | SYNPO2 | 0.788464 | 8 | 0 | 7 |
| DES | VMAC | 0.787526 | 3 | 0 | 3 |
| DES | LONRF2 | 0.785312 | 5 | 0 | 5 |
For details and further investigation, click here