| Full name: microtubule associated protein 7 | Alias Symbol: E-MAP-115 | ||
| Type: protein-coding gene | Cytoband: 6q23.3 | ||
| Entrez ID: 9053 | HGNC ID: HGNC:6869 | Ensembl Gene: ENSG00000135525 | OMIM ID: 604108 |
| Drug and gene relationship at DGIdb | |||
Expression of MAP7:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MAP7 | 9053 | 202890_at | 0.0020 | 0.9987 | |
| GSE20347 | MAP7 | 9053 | 202890_at | -0.4580 | 0.0289 | |
| GSE23400 | MAP7 | 9053 | 202889_x_at | -0.1599 | 0.0309 | |
| GSE26886 | MAP7 | 9053 | 202890_at | -0.6151 | 0.0168 | |
| GSE29001 | MAP7 | 9053 | 202890_at | -0.4194 | 0.1292 | |
| GSE38129 | MAP7 | 9053 | 202890_at | 0.0901 | 0.8215 | |
| GSE45670 | MAP7 | 9053 | 202890_at | 0.0379 | 0.8452 | |
| GSE53622 | MAP7 | 9053 | 71730 | 0.1181 | 0.5322 | |
| GSE53624 | MAP7 | 9053 | 71730 | -0.3714 | 0.0003 | |
| GSE63941 | MAP7 | 9053 | 202890_at | 2.4303 | 0.0005 | |
| GSE77861 | MAP7 | 9053 | 202890_at | -0.2616 | 0.4217 | |
| GSE97050 | MAP7 | 9053 | A_32_P190416 | 0.7513 | 0.2883 | |
| SRP007169 | MAP7 | 9053 | RNAseq | 0.9343 | 0.0445 | |
| SRP008496 | MAP7 | 9053 | RNAseq | 0.7326 | 0.0362 | |
| SRP064894 | MAP7 | 9053 | RNAseq | -0.5404 | 0.0826 | |
| SRP133303 | MAP7 | 9053 | RNAseq | -0.1208 | 0.5742 | |
| SRP159526 | MAP7 | 9053 | RNAseq | -0.4050 | 0.1952 | |
| SRP193095 | MAP7 | 9053 | RNAseq | -0.6363 | 0.0000 | |
| SRP219564 | MAP7 | 9053 | RNAseq | -0.0880 | 0.9177 | |
| TCGA | MAP7 | 9053 | RNAseq | 0.1210 | 0.2033 |
Upregulated datasets: 1; Downregulated datasets: 0.
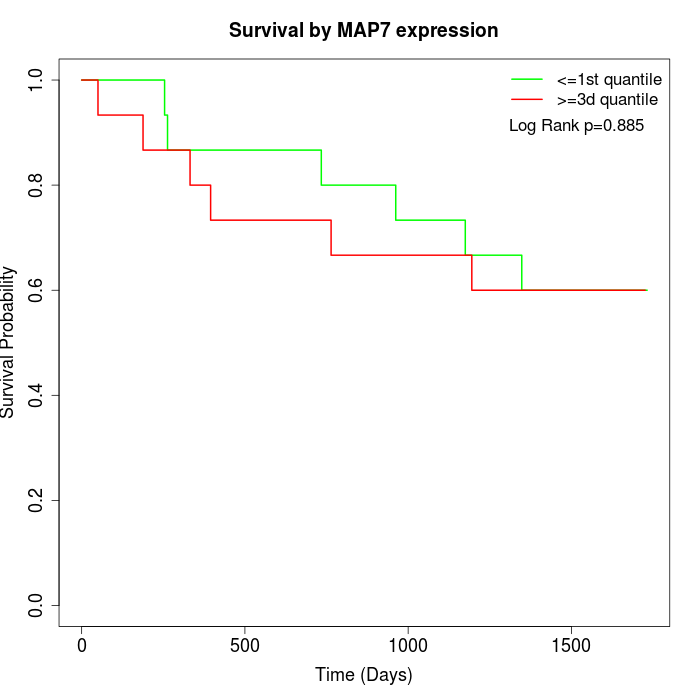
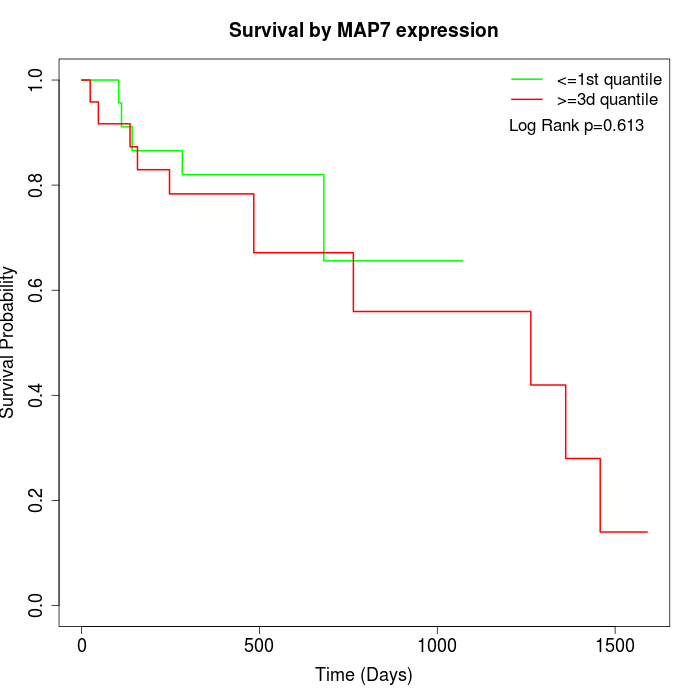
Survival by MAP7 expression:
Note: Click image to view full size file.
Copy number change of MAP7:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MAP7 | 9053 | 3 | 5 | 22 | |
| GSE20123 | MAP7 | 9053 | 3 | 3 | 24 | |
| GSE43470 | MAP7 | 9053 | 5 | 0 | 38 | |
| GSE46452 | MAP7 | 9053 | 3 | 10 | 46 | |
| GSE47630 | MAP7 | 9053 | 9 | 4 | 27 | |
| GSE54993 | MAP7 | 9053 | 3 | 2 | 65 | |
| GSE54994 | MAP7 | 9053 | 9 | 7 | 37 | |
| GSE60625 | MAP7 | 9053 | 0 | 1 | 10 | |
| GSE74703 | MAP7 | 9053 | 4 | 0 | 32 | |
| GSE74704 | MAP7 | 9053 | 0 | 2 | 18 | |
| TCGA | MAP7 | 9053 | 11 | 19 | 66 |
Total number of gains: 50; Total number of losses: 53; Total Number of normals: 385.
Somatic mutations of MAP7:
Generating mutation plots.
Highly correlated genes for MAP7:
Showing top 20/665 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MAP7 | SPINT2 | 0.780358 | 5 | 0 | 5 |
| MAP7 | FAM83H | 0.769584 | 4 | 0 | 4 |
| MAP7 | PSENEN | 0.769087 | 3 | 0 | 3 |
| MAP7 | CYB561 | 0.737269 | 4 | 0 | 4 |
| MAP7 | AP2S1 | 0.730179 | 3 | 0 | 3 |
| MAP7 | AP1S3 | 0.725156 | 5 | 0 | 4 |
| MAP7 | SRD5A1 | 0.723387 | 4 | 0 | 4 |
| MAP7 | FAM110A | 0.720746 | 4 | 0 | 4 |
| MAP7 | STON2 | 0.712208 | 3 | 0 | 3 |
| MAP7 | MFAP3 | 0.70682 | 4 | 0 | 4 |
| MAP7 | SLC25A39 | 0.69557 | 5 | 0 | 4 |
| MAP7 | GJB3 | 0.690865 | 7 | 0 | 6 |
| MAP7 | SYTL1 | 0.690057 | 5 | 0 | 4 |
| MAP7 | B4GALNT3 | 0.686505 | 3 | 0 | 3 |
| MAP7 | CHMP4C | 0.686052 | 7 | 0 | 5 |
| MAP7 | RNF149 | 0.685955 | 4 | 0 | 4 |
| MAP7 | TADA1 | 0.683077 | 3 | 0 | 3 |
| MAP7 | MARVELD2 | 0.6825 | 7 | 0 | 5 |
| MAP7 | TMX1 | 0.679927 | 4 | 0 | 4 |
| MAP7 | CXADR | 0.675863 | 8 | 0 | 6 |
For details and further investigation, click here