| Full name: MAP7 domain containing 1 | Alias Symbol: FLJ10350|FLJ39022 | ||
| Type: protein-coding gene | Cytoband: 1p34.3 | ||
| Entrez ID: 55700 | HGNC ID: HGNC:25514 | Ensembl Gene: ENSG00000116871 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of MAP7D1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MAP7D1 | 55700 | 217943_s_at | 0.4273 | 0.3619 | |
| GSE20347 | MAP7D1 | 55700 | 217943_s_at | 0.1071 | 0.6592 | |
| GSE23400 | MAP7D1 | 55700 | 217943_s_at | 0.2211 | 0.0137 | |
| GSE26886 | MAP7D1 | 55700 | 217943_s_at | -0.2244 | 0.3437 | |
| GSE29001 | MAP7D1 | 55700 | 217943_s_at | 0.1334 | 0.6679 | |
| GSE38129 | MAP7D1 | 55700 | 217943_s_at | 0.0148 | 0.9568 | |
| GSE45670 | MAP7D1 | 55700 | 217943_s_at | 0.5636 | 0.0240 | |
| GSE53622 | MAP7D1 | 55700 | 154517 | 0.2452 | 0.0166 | |
| GSE53624 | MAP7D1 | 55700 | 14856 | 0.3264 | 0.0000 | |
| GSE63941 | MAP7D1 | 55700 | 217943_s_at | -1.4367 | 0.0066 | |
| GSE77861 | MAP7D1 | 55700 | 217943_s_at | 0.7624 | 0.0546 | |
| GSE97050 | MAP7D1 | 55700 | A_33_P3380101 | -0.0643 | 0.7432 | |
| SRP007169 | MAP7D1 | 55700 | RNAseq | 0.6290 | 0.2394 | |
| SRP008496 | MAP7D1 | 55700 | RNAseq | 0.7210 | 0.0693 | |
| SRP064894 | MAP7D1 | 55700 | RNAseq | 0.3754 | 0.0644 | |
| SRP133303 | MAP7D1 | 55700 | RNAseq | 0.6184 | 0.0147 | |
| SRP159526 | MAP7D1 | 55700 | RNAseq | -0.3239 | 0.3420 | |
| SRP193095 | MAP7D1 | 55700 | RNAseq | 1.0676 | 0.0000 | |
| SRP219564 | MAP7D1 | 55700 | RNAseq | -0.2637 | 0.5844 | |
| TCGA | MAP7D1 | 55700 | RNAseq | 0.1165 | 0.0645 |
Upregulated datasets: 1; Downregulated datasets: 1.
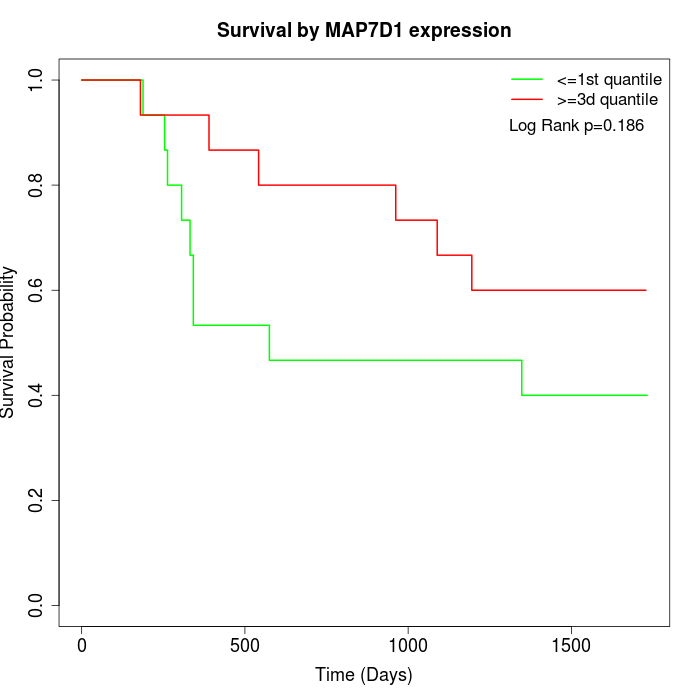
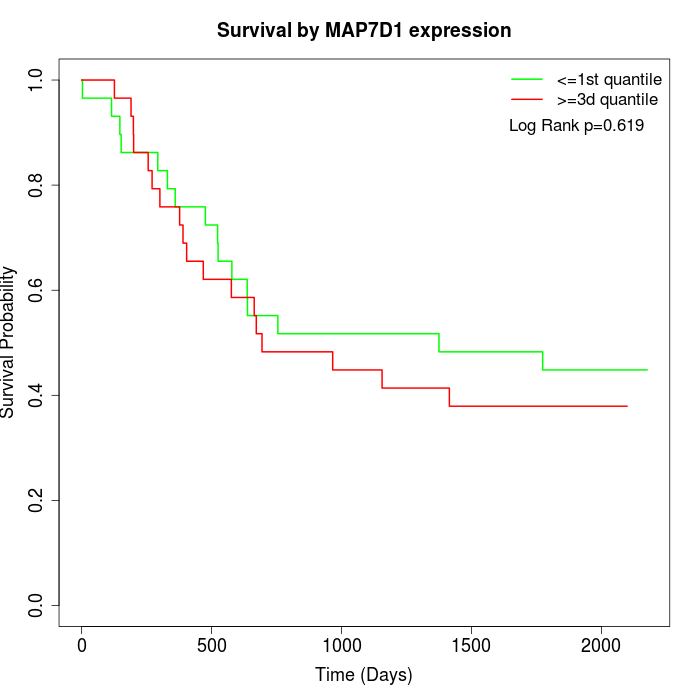
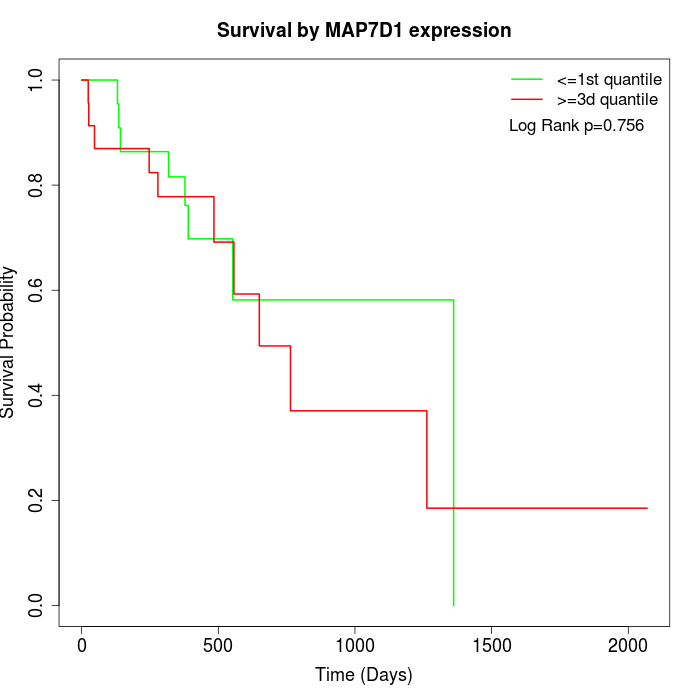
Survival by MAP7D1 expression:
Note: Click image to view full size file.
Copy number change of MAP7D1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MAP7D1 | 55700 | 3 | 5 | 22 | |
| GSE20123 | MAP7D1 | 55700 | 3 | 4 | 23 | |
| GSE43470 | MAP7D1 | 55700 | 7 | 2 | 34 | |
| GSE46452 | MAP7D1 | 55700 | 4 | 1 | 54 | |
| GSE47630 | MAP7D1 | 55700 | 8 | 3 | 29 | |
| GSE54993 | MAP7D1 | 55700 | 1 | 1 | 68 | |
| GSE54994 | MAP7D1 | 55700 | 11 | 2 | 40 | |
| GSE60625 | MAP7D1 | 55700 | 0 | 0 | 11 | |
| GSE74703 | MAP7D1 | 55700 | 6 | 1 | 29 | |
| GSE74704 | MAP7D1 | 55700 | 2 | 0 | 18 | |
| TCGA | MAP7D1 | 55700 | 11 | 19 | 66 |
Total number of gains: 56; Total number of losses: 38; Total Number of normals: 394.
Somatic mutations of MAP7D1:
Generating mutation plots.
Highly correlated genes for MAP7D1:
Showing top 20/303 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MAP7D1 | CLASP1 | 0.712446 | 3 | 0 | 3 |
| MAP7D1 | TSSC4 | 0.694311 | 3 | 0 | 3 |
| MAP7D1 | SPRY4-IT1 | 0.691919 | 3 | 0 | 3 |
| MAP7D1 | SLC39A1 | 0.685174 | 4 | 0 | 4 |
| MAP7D1 | TMCC2 | 0.665945 | 3 | 0 | 3 |
| MAP7D1 | LINC01137 | 0.664267 | 5 | 0 | 4 |
| MAP7D1 | STK17A | 0.657228 | 5 | 0 | 4 |
| MAP7D1 | FBLN2 | 0.654474 | 4 | 0 | 4 |
| MAP7D1 | SH2D5 | 0.641186 | 8 | 0 | 6 |
| MAP7D1 | LAMB2 | 0.637517 | 3 | 0 | 3 |
| MAP7D1 | LRRC32 | 0.637399 | 3 | 0 | 3 |
| MAP7D1 | TTYH3 | 0.635854 | 4 | 0 | 3 |
| MAP7D1 | OSR1 | 0.632876 | 3 | 0 | 3 |
| MAP7D1 | STRIP2 | 0.632133 | 4 | 0 | 4 |
| MAP7D1 | BAHCC1 | 0.630667 | 4 | 0 | 3 |
| MAP7D1 | CTPS1 | 0.627927 | 6 | 0 | 5 |
| MAP7D1 | MLXIP | 0.627393 | 7 | 0 | 6 |
| MAP7D1 | MMP14 | 0.625024 | 5 | 0 | 4 |
| MAP7D1 | POFUT2 | 0.62254 | 4 | 0 | 3 |
| MAP7D1 | C19orf12 | 0.621927 | 4 | 0 | 3 |
For details and further investigation, click here