| Full name: metallothionein 3 | Alias Symbol: GIF | ||
| Type: protein-coding gene | Cytoband: 16q13 | ||
| Entrez ID: 4504 | HGNC ID: HGNC:7408 | Ensembl Gene: ENSG00000087250 | OMIM ID: 139255 |
| Drug and gene relationship at DGIdb | |||
Expression of MT3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MT3 | 4504 | 205970_at | -0.1050 | 0.6865 | |
| GSE20347 | MT3 | 4504 | 205970_at | -0.1030 | 0.1906 | |
| GSE23400 | MT3 | 4504 | 205970_at | -0.1231 | 0.0038 | |
| GSE26886 | MT3 | 4504 | 205970_at | -0.0167 | 0.9401 | |
| GSE29001 | MT3 | 4504 | 205970_at | -0.0723 | 0.6404 | |
| GSE38129 | MT3 | 4504 | 205970_at | -0.2578 | 0.0135 | |
| GSE45670 | MT3 | 4504 | 205970_at | 0.0778 | 0.4058 | |
| GSE63941 | MT3 | 4504 | 205970_at | -0.1608 | 0.4688 | |
| GSE77861 | MT3 | 4504 | 205970_at | -0.2120 | 0.0376 | |
| GSE97050 | MT3 | 4504 | A_23_P129629 | -0.2153 | 0.3835 | |
| TCGA | MT3 | 4504 | RNAseq | -2.5343 | 0.0024 |
Upregulated datasets: 0; Downregulated datasets: 1.
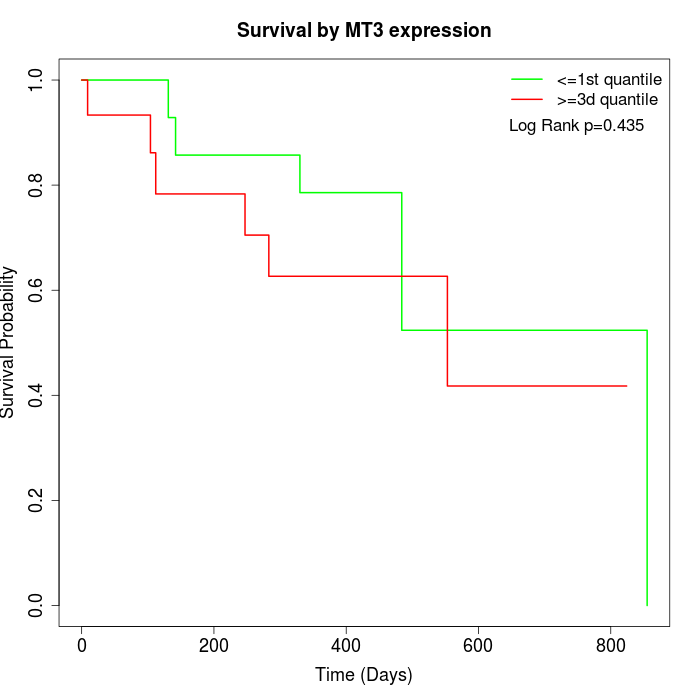
Survival by MT3 expression:
Note: Click image to view full size file.
Copy number change of MT3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MT3 | 4504 | 5 | 0 | 25 | |
| GSE20123 | MT3 | 4504 | 5 | 1 | 24 | |
| GSE43470 | MT3 | 4504 | 2 | 7 | 34 | |
| GSE46452 | MT3 | 4504 | 38 | 1 | 20 | |
| GSE47630 | MT3 | 4504 | 10 | 8 | 22 | |
| GSE54993 | MT3 | 4504 | 2 | 4 | 64 | |
| GSE54994 | MT3 | 4504 | 7 | 10 | 36 | |
| GSE60625 | MT3 | 4504 | 4 | 0 | 7 | |
| GSE74703 | MT3 | 4504 | 2 | 4 | 30 | |
| GSE74704 | MT3 | 4504 | 4 | 0 | 16 | |
| TCGA | MT3 | 4504 | 25 | 12 | 59 |
Total number of gains: 104; Total number of losses: 47; Total Number of normals: 337.
Somatic mutations of MT3:
Generating mutation plots.
Highly correlated genes for MT3:
Showing top 20/582 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MT3 | CACNG8 | 0.737055 | 3 | 0 | 3 |
| MT3 | CCDC157 | 0.708884 | 3 | 0 | 3 |
| MT3 | SUN5 | 0.708085 | 3 | 0 | 3 |
| MT3 | SIM1 | 0.705288 | 4 | 0 | 3 |
| MT3 | KLHL10 | 0.704141 | 4 | 0 | 4 |
| MT3 | SLC25A47 | 0.70035 | 3 | 0 | 3 |
| MT3 | SLC45A2 | 0.699793 | 4 | 0 | 4 |
| MT3 | UPB1 | 0.692753 | 4 | 0 | 4 |
| MT3 | CPN1 | 0.689495 | 5 | 0 | 5 |
| MT3 | OPN4 | 0.689246 | 3 | 0 | 3 |
| MT3 | SH2D1B | 0.68212 | 3 | 0 | 3 |
| MT3 | CELF5 | 0.677233 | 4 | 0 | 4 |
| MT3 | PRX | 0.675741 | 4 | 0 | 3 |
| MT3 | CALCA | 0.674979 | 5 | 0 | 5 |
| MT3 | PGAM2 | 0.672146 | 3 | 0 | 3 |
| MT3 | TRH | 0.668697 | 5 | 0 | 4 |
| MT3 | CA4 | 0.667714 | 7 | 0 | 6 |
| MT3 | GRK4 | 0.665791 | 4 | 0 | 4 |
| MT3 | FCN2 | 0.665478 | 4 | 0 | 4 |
| MT3 | GRM4 | 0.661758 | 5 | 0 | 5 |
For details and further investigation, click here