| Full name: myosin light chain 12B | Alias Symbol: MRLC2 | ||
| Type: protein-coding gene | Cytoband: 18p11.31 | ||
| Entrez ID: 103910 | HGNC ID: HGNC:29827 | Ensembl Gene: ENSG00000118680 | OMIM ID: 609211 |
| Drug and gene relationship at DGIdb | |||
MYL12B involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04510 | Focal adhesion | |
| hsa04530 | Tight junction | |
| hsa04611 | Platelet activation | |
| hsa04670 | Leukocyte transendothelial migration | |
| hsa04810 | Regulation of actin cytoskeleton |
Expression of MYL12B:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MYL12B | 103910 | 221474_at | -0.1123 | 0.8628 | |
| GSE20347 | MYL12B | 103910 | 221474_at | -0.3094 | 0.0191 | |
| GSE23400 | MYL12B | 103910 | 221474_at | -0.0594 | 0.4339 | |
| GSE26886 | MYL12B | 103910 | 221474_at | -0.3359 | 0.0381 | |
| GSE29001 | MYL12B | 103910 | 221474_at | -0.2691 | 0.0608 | |
| GSE38129 | MYL12B | 103910 | 221474_at | -0.1280 | 0.3737 | |
| GSE45670 | MYL12B | 103910 | 221474_at | -0.0490 | 0.7271 | |
| GSE53622 | MYL12B | 103910 | 140901 | -0.6475 | 0.0000 | |
| GSE53624 | MYL12B | 103910 | 140901 | -0.1888 | 0.0034 | |
| GSE63941 | MYL12B | 103910 | 221474_at | 0.1439 | 0.7818 | |
| GSE77861 | MYL12B | 103910 | 221474_at | -0.1165 | 0.7385 | |
| GSE97050 | MYL12B | 103910 | A_24_P75230 | 0.1888 | 0.5008 | |
| SRP007169 | MYL12B | 103910 | RNAseq | -1.4379 | 0.0003 | |
| SRP008496 | MYL12B | 103910 | RNAseq | -1.3141 | 0.0000 | |
| SRP064894 | MYL12B | 103910 | RNAseq | 0.5309 | 0.1132 | |
| SRP133303 | MYL12B | 103910 | RNAseq | -0.1728 | 0.3184 | |
| SRP159526 | MYL12B | 103910 | RNAseq | -0.4119 | 0.0806 | |
| SRP193095 | MYL12B | 103910 | RNAseq | -0.3722 | 0.0451 | |
| SRP219564 | MYL12B | 103910 | RNAseq | -0.7439 | 0.0685 | |
| TCGA | MYL12B | 103910 | RNAseq | 0.0834 | 0.1861 |
Upregulated datasets: 0; Downregulated datasets: 2.
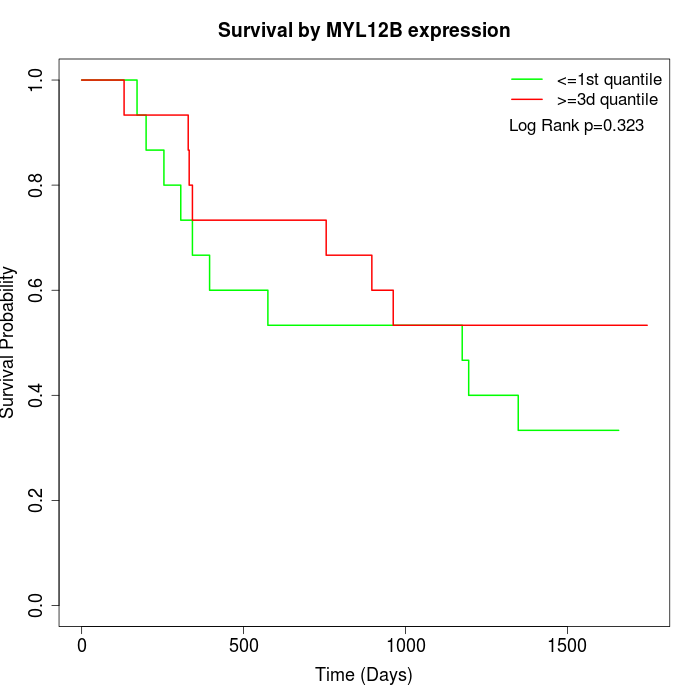
Survival by MYL12B expression:
Note: Click image to view full size file.
Copy number change of MYL12B:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MYL12B | 103910 | 8 | 1 | 21 | |
| GSE20123 | MYL12B | 103910 | 8 | 1 | 21 | |
| GSE43470 | MYL12B | 103910 | 4 | 5 | 34 | |
| GSE46452 | MYL12B | 103910 | 3 | 20 | 36 | |
| GSE47630 | MYL12B | 103910 | 8 | 17 | 15 | |
| GSE54993 | MYL12B | 103910 | 6 | 4 | 60 | |
| GSE54994 | MYL12B | 103910 | 11 | 8 | 34 | |
| GSE60625 | MYL12B | 103910 | 0 | 4 | 7 | |
| GSE74703 | MYL12B | 103910 | 3 | 3 | 30 | |
| GSE74704 | MYL12B | 103910 | 6 | 1 | 13 | |
| TCGA | MYL12B | 103910 | 35 | 19 | 42 |
Total number of gains: 92; Total number of losses: 83; Total Number of normals: 313.
Somatic mutations of MYL12B:
Generating mutation plots.
Highly correlated genes for MYL12B:
Showing top 20/531 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MYL12B | MYL12A | 0.815629 | 12 | 0 | 11 |
| MYL12B | PTGR2 | 0.752439 | 3 | 0 | 3 |
| MYL12B | ZCCHC10 | 0.740272 | 3 | 0 | 3 |
| MYL12B | RNF183 | 0.736168 | 3 | 0 | 3 |
| MYL12B | SS18 | 0.733317 | 3 | 0 | 3 |
| MYL12B | WDFY2 | 0.703431 | 3 | 0 | 3 |
| MYL12B | AP1S3 | 0.698388 | 4 | 0 | 3 |
| MYL12B | STXBP5 | 0.695242 | 3 | 0 | 3 |
| MYL12B | LRRC40 | 0.693538 | 3 | 0 | 3 |
| MYL12B | HEATR5B | 0.686586 | 3 | 0 | 3 |
| MYL12B | TMEM19 | 0.68635 | 4 | 0 | 3 |
| MYL12B | PPP4R1 | 0.686228 | 11 | 0 | 10 |
| MYL12B | PGGT1B | 0.683484 | 3 | 0 | 3 |
| MYL12B | VPS25 | 0.682923 | 3 | 0 | 3 |
| MYL12B | ZNF823 | 0.682021 | 3 | 0 | 3 |
| MYL12B | SRD5A3 | 0.680541 | 3 | 0 | 3 |
| MYL12B | AGA | 0.678204 | 3 | 0 | 3 |
| MYL12B | DUSP18 | 0.670285 | 4 | 0 | 3 |
| MYL12B | UNC13D | 0.652251 | 4 | 0 | 4 |
| MYL12B | RNF144B | 0.651442 | 4 | 0 | 4 |
For details and further investigation, click here