| Full name: neutrophil cytosolic factor 2 | Alias Symbol: p67phox|NOXA2 | ||
| Type: protein-coding gene | Cytoband: 1q25.3 | ||
| Entrez ID: 4688 | HGNC ID: HGNC:7661 | Ensembl Gene: ENSG00000116701 | OMIM ID: 608515 |
| Drug and gene relationship at DGIdb | |||
NCF2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04380 | Osteoclast differentiation | |
| hsa04670 | Leukocyte transendothelial migration | |
| hsa05140 | Leishmaniasis |
Expression of NCF2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NCF2 | 4688 | 209949_at | 1.4604 | 0.1823 | |
| GSE20347 | NCF2 | 4688 | 209949_at | 1.3488 | 0.0003 | |
| GSE23400 | NCF2 | 4688 | 209949_at | 0.6078 | 0.0000 | |
| GSE26886 | NCF2 | 4688 | 209949_at | 0.0906 | 0.8635 | |
| GSE29001 | NCF2 | 4688 | 209949_at | 0.8926 | 0.0054 | |
| GSE38129 | NCF2 | 4688 | 209949_at | 1.4070 | 0.0000 | |
| GSE45670 | NCF2 | 4688 | 209949_at | 1.4221 | 0.0031 | |
| GSE53622 | NCF2 | 4688 | 32038 | 0.8901 | 0.0000 | |
| GSE53624 | NCF2 | 4688 | 32038 | 1.3046 | 0.0000 | |
| GSE63941 | NCF2 | 4688 | 209949_at | 0.7738 | 0.4429 | |
| GSE77861 | NCF2 | 4688 | 209949_at | 1.3758 | 0.0176 | |
| GSE97050 | NCF2 | 4688 | A_23_P138194 | 1.0937 | 0.0620 | |
| SRP007169 | NCF2 | 4688 | RNAseq | 2.3925 | 0.0003 | |
| SRP008496 | NCF2 | 4688 | RNAseq | 1.4404 | 0.0016 | |
| SRP064894 | NCF2 | 4688 | RNAseq | 1.2460 | 0.0000 | |
| SRP133303 | NCF2 | 4688 | RNAseq | 1.3725 | 0.0000 | |
| SRP159526 | NCF2 | 4688 | RNAseq | 1.3510 | 0.0865 | |
| SRP193095 | NCF2 | 4688 | RNAseq | 1.8452 | 0.0000 | |
| SRP219564 | NCF2 | 4688 | RNAseq | 1.5720 | 0.0301 | |
| TCGA | NCF2 | 4688 | RNAseq | 0.9210 | 0.0000 |
Upregulated datasets: 11; Downregulated datasets: 0.
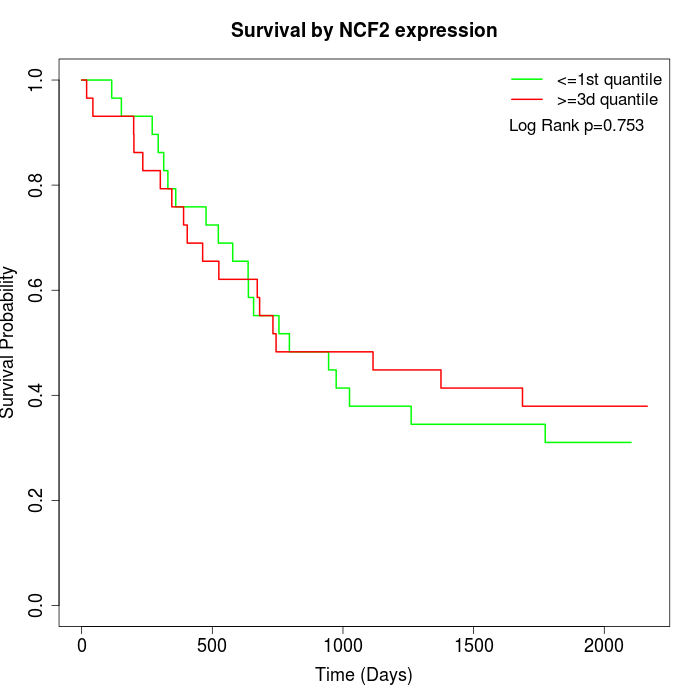
Survival by NCF2 expression:
Note: Click image to view full size file.
Copy number change of NCF2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NCF2 | 4688 | 12 | 0 | 18 | |
| GSE20123 | NCF2 | 4688 | 12 | 0 | 18 | |
| GSE43470 | NCF2 | 4688 | 7 | 1 | 35 | |
| GSE46452 | NCF2 | 4688 | 3 | 1 | 55 | |
| GSE47630 | NCF2 | 4688 | 14 | 0 | 26 | |
| GSE54993 | NCF2 | 4688 | 0 | 6 | 64 | |
| GSE54994 | NCF2 | 4688 | 14 | 1 | 38 | |
| GSE60625 | NCF2 | 4688 | 0 | 0 | 11 | |
| GSE74703 | NCF2 | 4688 | 7 | 1 | 28 | |
| GSE74704 | NCF2 | 4688 | 5 | 0 | 15 | |
| TCGA | NCF2 | 4688 | 40 | 3 | 53 |
Total number of gains: 114; Total number of losses: 13; Total Number of normals: 361.
Somatic mutations of NCF2:
Generating mutation plots.
Highly correlated genes for NCF2:
Showing top 20/1435 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NCF2 | ALPK2 | 0.796546 | 3 | 0 | 3 |
| NCF2 | KTI12 | 0.781836 | 3 | 0 | 3 |
| NCF2 | LRRC8C | 0.770004 | 3 | 0 | 3 |
| NCF2 | ALG10 | 0.766392 | 3 | 0 | 3 |
| NCF2 | ZNF469 | 0.766143 | 5 | 0 | 5 |
| NCF2 | FCGR3A | 0.75612 | 3 | 0 | 3 |
| NCF2 | MOB3C | 0.746904 | 3 | 0 | 3 |
| NCF2 | PPHLN1 | 0.729535 | 3 | 0 | 3 |
| NCF2 | C19orf38 | 0.728954 | 3 | 0 | 3 |
| NCF2 | VPS35 | 0.728056 | 3 | 0 | 3 |
| NCF2 | C9orf78 | 0.727697 | 3 | 0 | 3 |
| NCF2 | ALKBH6 | 0.725382 | 3 | 0 | 3 |
| NCF2 | KIF13A | 0.722379 | 3 | 0 | 3 |
| NCF2 | XPR1 | 0.719994 | 6 | 0 | 5 |
| NCF2 | IKBIP | 0.716471 | 6 | 0 | 6 |
| NCF2 | IRF2BPL | 0.714579 | 3 | 0 | 3 |
| NCF2 | COMMD7 | 0.713777 | 3 | 0 | 3 |
| NCF2 | WDR54 | 0.71293 | 6 | 0 | 5 |
| NCF2 | LRRC58 | 0.707707 | 3 | 0 | 3 |
| NCF2 | MB21D2 | 0.705917 | 6 | 0 | 6 |
For details and further investigation, click here