| Full name: nuclear receptor coactivator 3 | Alias Symbol: RAC3|AIB1|ACTR|p/CIP|TRAM-1|CAGH16|TNRC16|KAT13B|bHLHe42|SRC-3|SRC3 | ||
| Type: protein-coding gene | Cytoband: 20q13.12 | ||
| Entrez ID: 8202 | HGNC ID: HGNC:7670 | Ensembl Gene: ENSG00000124151 | OMIM ID: 601937 |
| Drug and gene relationship at DGIdb | |||
Expression of NCOA3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NCOA3 | 8202 | 207700_s_at | 0.4027 | 0.5092 | |
| GSE20347 | NCOA3 | 8202 | 207700_s_at | -0.2480 | 0.2772 | |
| GSE23400 | NCOA3 | 8202 | 207700_s_at | 0.1490 | 0.2300 | |
| GSE26886 | NCOA3 | 8202 | 209061_at | -0.3689 | 0.2983 | |
| GSE29001 | NCOA3 | 8202 | 207700_s_at | 0.1911 | 0.6148 | |
| GSE38129 | NCOA3 | 8202 | 207700_s_at | -0.0671 | 0.7672 | |
| GSE45670 | NCOA3 | 8202 | 207700_s_at | 0.2767 | 0.3066 | |
| GSE53622 | NCOA3 | 8202 | 8105 | 0.3869 | 0.0000 | |
| GSE53624 | NCOA3 | 8202 | 8105 | 0.2642 | 0.0000 | |
| GSE63941 | NCOA3 | 8202 | 207700_s_at | 0.4584 | 0.5454 | |
| GSE77861 | NCOA3 | 8202 | 209061_at | 0.1617 | 0.3716 | |
| GSE97050 | NCOA3 | 8202 | A_23_P120442 | 0.0783 | 0.7698 | |
| SRP007169 | NCOA3 | 8202 | RNAseq | -0.1156 | 0.7714 | |
| SRP008496 | NCOA3 | 8202 | RNAseq | 0.1054 | 0.6698 | |
| SRP064894 | NCOA3 | 8202 | RNAseq | 0.0439 | 0.8486 | |
| SRP133303 | NCOA3 | 8202 | RNAseq | 0.2340 | 0.1526 | |
| SRP159526 | NCOA3 | 8202 | RNAseq | 0.1176 | 0.8291 | |
| SRP193095 | NCOA3 | 8202 | RNAseq | 0.1438 | 0.2213 | |
| SRP219564 | NCOA3 | 8202 | RNAseq | -0.0194 | 0.9507 | |
| TCGA | NCOA3 | 8202 | RNAseq | 0.1254 | 0.0216 |
Upregulated datasets: 0; Downregulated datasets: 0.
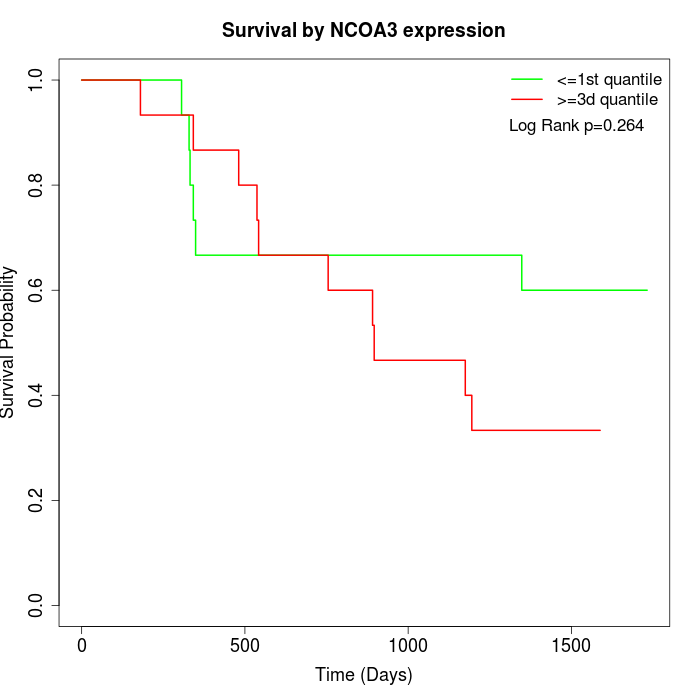
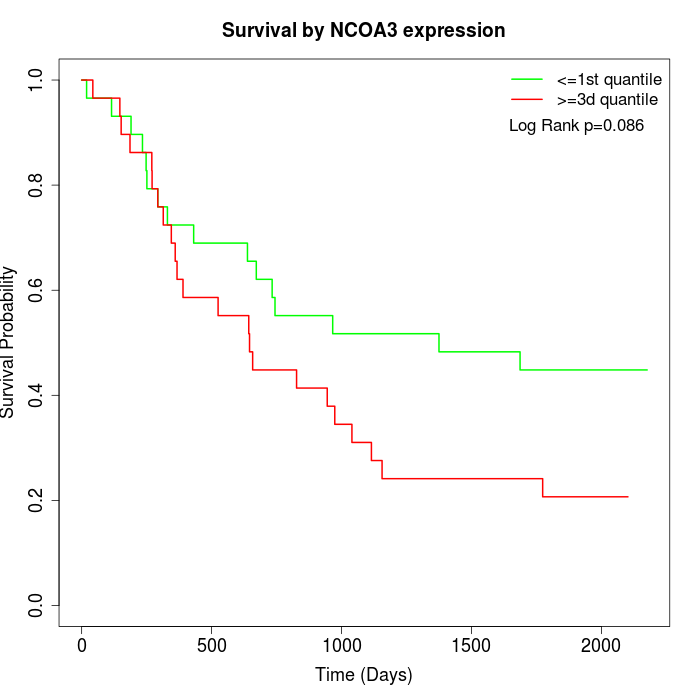
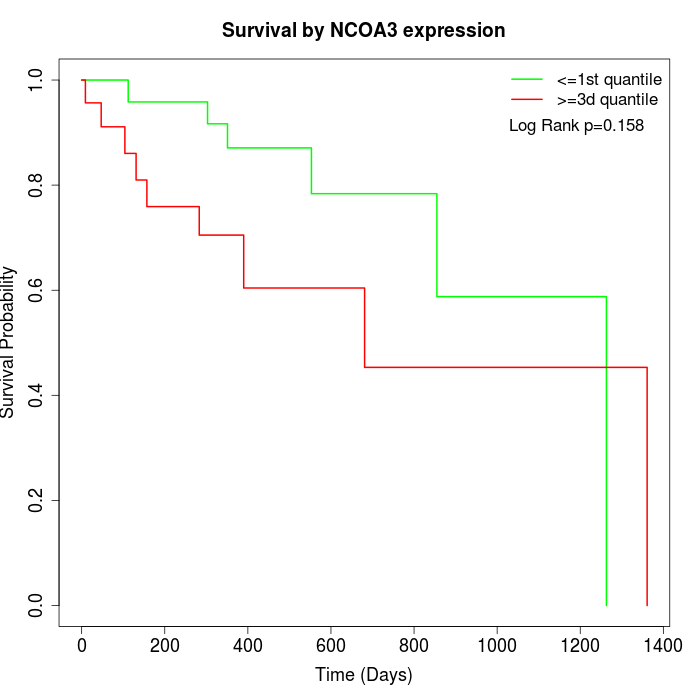
Survival by NCOA3 expression:
Note: Click image to view full size file.
Copy number change of NCOA3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NCOA3 | 8202 | 15 | 2 | 13 | |
| GSE20123 | NCOA3 | 8202 | 15 | 2 | 13 | |
| GSE43470 | NCOA3 | 8202 | 12 | 1 | 30 | |
| GSE46452 | NCOA3 | 8202 | 29 | 0 | 30 | |
| GSE47630 | NCOA3 | 8202 | 24 | 1 | 15 | |
| GSE54993 | NCOA3 | 8202 | 0 | 18 | 52 | |
| GSE54994 | NCOA3 | 8202 | 26 | 0 | 27 | |
| GSE60625 | NCOA3 | 8202 | 0 | 0 | 11 | |
| GSE74703 | NCOA3 | 8202 | 10 | 1 | 25 | |
| GSE74704 | NCOA3 | 8202 | 11 | 0 | 9 | |
| TCGA | NCOA3 | 8202 | 45 | 3 | 48 |
Total number of gains: 187; Total number of losses: 28; Total Number of normals: 273.
Somatic mutations of NCOA3:
Generating mutation plots.
Highly correlated genes for NCOA3:
Showing top 20/192 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NCOA3 | PSMA2 | 0.765623 | 3 | 0 | 3 |
| NCOA3 | POLR3F | 0.751276 | 3 | 0 | 3 |
| NCOA3 | CCT6A | 0.740568 | 3 | 0 | 3 |
| NCOA3 | ATP7A | 0.721044 | 3 | 0 | 3 |
| NCOA3 | TBC1D23 | 0.712207 | 3 | 0 | 3 |
| NCOA3 | HNRNPF | 0.71069 | 3 | 0 | 3 |
| NCOA3 | ZCCHC8 | 0.699493 | 3 | 0 | 3 |
| NCOA3 | HDAC3 | 0.6921 | 3 | 0 | 3 |
| NCOA3 | VTI1B | 0.687929 | 3 | 0 | 3 |
| NCOA3 | KATNA1 | 0.683138 | 3 | 0 | 3 |
| NCOA3 | NAA35 | 0.680666 | 3 | 0 | 3 |
| NCOA3 | HARS2 | 0.679306 | 4 | 0 | 4 |
| NCOA3 | TRAP1 | 0.678386 | 3 | 0 | 3 |
| NCOA3 | SPAG9 | 0.674513 | 4 | 0 | 4 |
| NCOA3 | IMPACT | 0.668179 | 3 | 0 | 3 |
| NCOA3 | ITGB5 | 0.665511 | 3 | 0 | 3 |
| NCOA3 | ANKZF1 | 0.660307 | 4 | 0 | 3 |
| NCOA3 | RBM27 | 0.65732 | 3 | 0 | 3 |
| NCOA3 | HSPA4 | 0.654554 | 5 | 0 | 3 |
| NCOA3 | C12orf65 | 0.652909 | 4 | 0 | 3 |
For details and further investigation, click here