| Full name: neurofibromin 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 17q11.2 | ||
| Entrez ID: 4763 | HGNC ID: HGNC:7765 | Ensembl Gene: ENSG00000196712 | OMIM ID: 613113 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
NF1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04010 | MAPK signaling pathway | |
| hsa04014 | Ras signaling pathway |
Expression of NF1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | NF1 | 4763 | 39537 | 0.7096 | 0.0000 | |
| GSE53624 | NF1 | 4763 | 3700 | 0.4810 | 0.0000 | |
| GSE97050 | NF1 | 4763 | A_24_P1919 | 0.5148 | 0.1512 | |
| SRP007169 | NF1 | 4763 | RNAseq | 1.8687 | 0.0000 | |
| SRP008496 | NF1 | 4763 | RNAseq | 1.6611 | 0.0000 | |
| SRP064894 | NF1 | 4763 | RNAseq | 0.5982 | 0.0087 | |
| SRP133303 | NF1 | 4763 | RNAseq | 0.7010 | 0.0000 | |
| SRP159526 | NF1 | 4763 | RNAseq | 0.7083 | 0.0013 | |
| SRP193095 | NF1 | 4763 | RNAseq | 0.6520 | 0.0000 | |
| SRP219564 | NF1 | 4763 | RNAseq | 0.2451 | 0.5369 | |
| TCGA | NF1 | 4763 | RNAseq | 0.0266 | 0.6316 |
Upregulated datasets: 2; Downregulated datasets: 0.
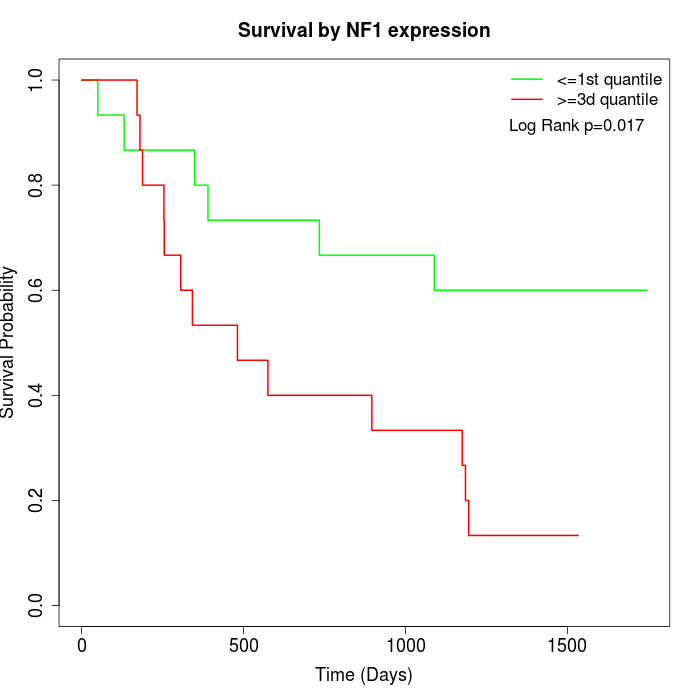
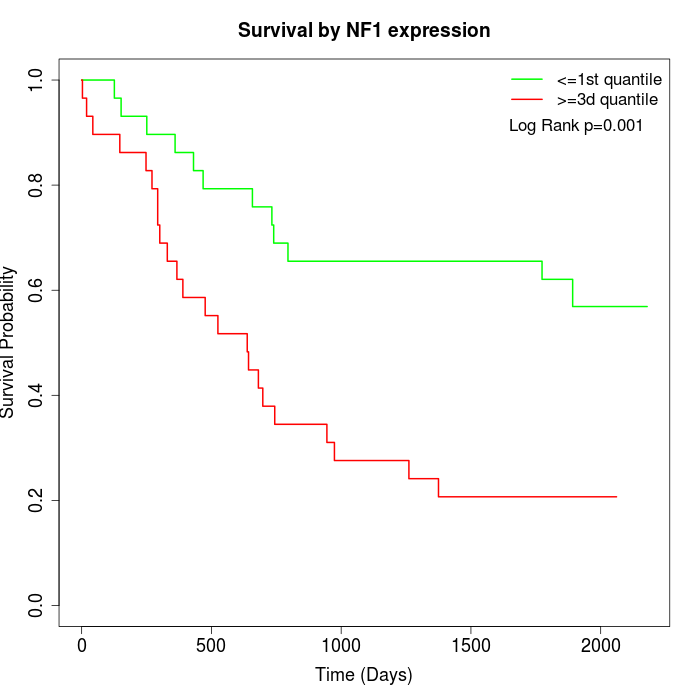
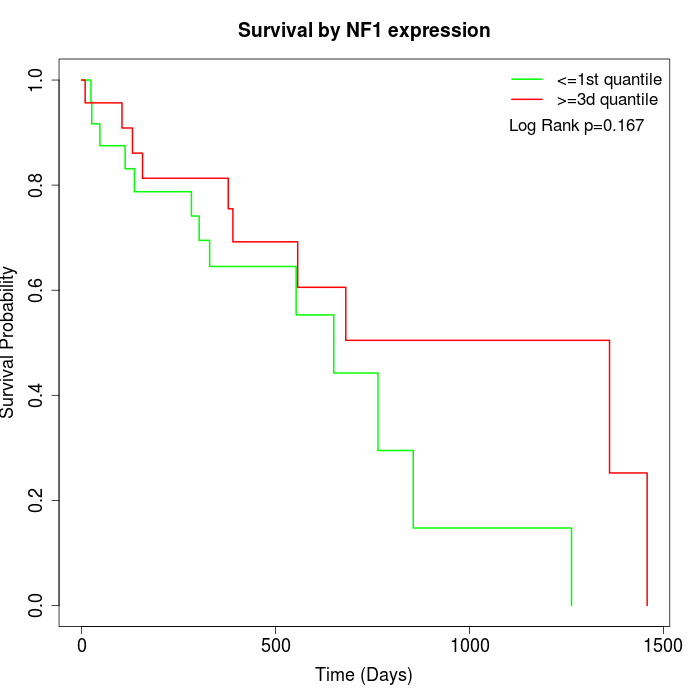
Survival by NF1 expression:
Note: Click image to view full size file.
Copy number change of NF1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NF1 | 4763 | 8 | 2 | 20 | |
| GSE20123 | NF1 | 4763 | 9 | 2 | 19 | |
| GSE43470 | NF1 | 4763 | 0 | 1 | 42 | |
| GSE46452 | NF1 | 4763 | 33 | 1 | 25 | |
| GSE47630 | NF1 | 4763 | 7 | 1 | 32 | |
| GSE54993 | NF1 | 4763 | 2 | 3 | 65 | |
| GSE54994 | NF1 | 4763 | 7 | 6 | 40 | |
| GSE60625 | NF1 | 4763 | 4 | 0 | 7 | |
| GSE74703 | NF1 | 4763 | 0 | 1 | 35 | |
| GSE74704 | NF1 | 4763 | 6 | 2 | 12 | |
| TCGA | NF1 | 4763 | 20 | 12 | 64 |
Total number of gains: 96; Total number of losses: 31; Total Number of normals: 361.
Somatic mutations of NF1:
Generating mutation plots.
Highly correlated genes for NF1:
Showing top 20/311 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NF1 | PRTFDC1 | 0.77558 | 3 | 0 | 3 |
| NF1 | RAI14 | 0.775282 | 3 | 0 | 3 |
| NF1 | ACVR1 | 0.774352 | 3 | 0 | 3 |
| NF1 | HIP1 | 0.772402 | 3 | 0 | 3 |
| NF1 | DHX57 | 0.77163 | 3 | 0 | 3 |
| NF1 | DEK | 0.768641 | 3 | 0 | 3 |
| NF1 | RAB34 | 0.767917 | 3 | 0 | 3 |
| NF1 | KIF3C | 0.766573 | 3 | 0 | 3 |
| NF1 | THOC5 | 0.76328 | 3 | 0 | 3 |
| NF1 | LPGAT1 | 0.762391 | 3 | 0 | 3 |
| NF1 | NEDD1 | 0.762334 | 3 | 0 | 3 |
| NF1 | CDC27 | 0.762184 | 3 | 0 | 3 |
| NF1 | MINPP1 | 0.760509 | 3 | 0 | 3 |
| NF1 | HMGN4 | 0.759376 | 3 | 0 | 3 |
| NF1 | GRAMD1A | 0.759296 | 3 | 0 | 3 |
| NF1 | WDR54 | 0.7587 | 3 | 0 | 3 |
| NF1 | AATF | 0.757368 | 3 | 0 | 3 |
| NF1 | TRIM37 | 0.75438 | 3 | 0 | 3 |
| NF1 | ZNF275 | 0.753274 | 3 | 0 | 3 |
| NF1 | ZNF281 | 0.753014 | 3 | 0 | 3 |
For details and further investigation, click here