| Full name: nuclear factor of activated T cells 4 | Alias Symbol: NFAT3 | ||
| Type: protein-coding gene | Cytoband: 14q12 | ||
| Entrez ID: 4776 | HGNC ID: HGNC:7778 | Ensembl Gene: ENSG00000100968 | OMIM ID: 602699 |
| Drug and gene relationship at DGIdb | |||
NFATC4 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04022 | cGMP-PKG signaling pathway | |
| hsa04310 | Wnt signaling pathway | |
| hsa04360 | Axon guidance | |
| hsa04921 | Oxytocin signaling pathway | |
| hsa05161 | Hepatitis B | |
| hsa05166 | HTLV-I infection |
Expression of NFATC4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NFATC4 | 4776 | 213345_at | 0.0089 | 0.9801 | |
| GSE20347 | NFATC4 | 4776 | 213345_at | 0.1002 | 0.2966 | |
| GSE23400 | NFATC4 | 4776 | 205897_at | -0.1976 | 0.0000 | |
| GSE26886 | NFATC4 | 4776 | 213345_at | 0.6289 | 0.0007 | |
| GSE29001 | NFATC4 | 4776 | 213345_at | 0.1064 | 0.7152 | |
| GSE38129 | NFATC4 | 4776 | 213345_at | -0.0455 | 0.5971 | |
| GSE45670 | NFATC4 | 4776 | 213345_at | -0.1903 | 0.0949 | |
| GSE53622 | NFATC4 | 4776 | 80708 | -1.0796 | 0.0000 | |
| GSE53624 | NFATC4 | 4776 | 80708 | -0.8833 | 0.0000 | |
| GSE63941 | NFATC4 | 4776 | 213345_at | -0.8476 | 0.0085 | |
| GSE77861 | NFATC4 | 4776 | 213345_at | -0.1905 | 0.3516 | |
| GSE97050 | NFATC4 | 4776 | A_33_P3250083 | -0.5471 | 0.1726 | |
| SRP007169 | NFATC4 | 4776 | RNAseq | 1.8011 | 0.0098 | |
| SRP064894 | NFATC4 | 4776 | RNAseq | 0.2164 | 0.3813 | |
| SRP133303 | NFATC4 | 4776 | RNAseq | -0.4373 | 0.1436 | |
| SRP159526 | NFATC4 | 4776 | RNAseq | 0.2409 | 0.5610 | |
| SRP193095 | NFATC4 | 4776 | RNAseq | 1.5253 | 0.0000 | |
| SRP219564 | NFATC4 | 4776 | RNAseq | -0.1318 | 0.8223 | |
| TCGA | NFATC4 | 4776 | RNAseq | -0.3576 | 0.0042 |
Upregulated datasets: 2; Downregulated datasets: 1.
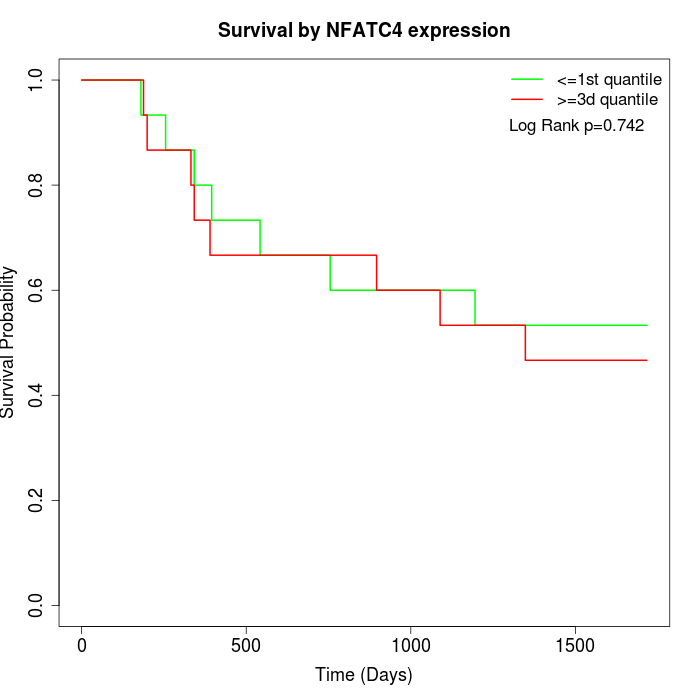
Survival by NFATC4 expression:
Note: Click image to view full size file.
Copy number change of NFATC4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NFATC4 | 4776 | 8 | 3 | 19 | |
| GSE20123 | NFATC4 | 4776 | 8 | 3 | 19 | |
| GSE43470 | NFATC4 | 4776 | 8 | 2 | 33 | |
| GSE46452 | NFATC4 | 4776 | 16 | 2 | 41 | |
| GSE47630 | NFATC4 | 4776 | 11 | 10 | 19 | |
| GSE54993 | NFATC4 | 4776 | 3 | 11 | 56 | |
| GSE54994 | NFATC4 | 4776 | 18 | 5 | 30 | |
| GSE60625 | NFATC4 | 4776 | 0 | 2 | 9 | |
| GSE74703 | NFATC4 | 4776 | 7 | 2 | 27 | |
| GSE74704 | NFATC4 | 4776 | 3 | 2 | 15 | |
| TCGA | NFATC4 | 4776 | 27 | 12 | 57 |
Total number of gains: 109; Total number of losses: 54; Total Number of normals: 325.
Somatic mutations of NFATC4:
Generating mutation plots.
Highly correlated genes for NFATC4:
Showing top 20/727 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NFATC4 | TNFSF12 | 0.84474 | 3 | 0 | 3 |
| NFATC4 | TCEAL1 | 0.831512 | 3 | 0 | 3 |
| NFATC4 | AKAP6 | 0.777431 | 3 | 0 | 3 |
| NFATC4 | CLEC3B | 0.774382 | 3 | 0 | 3 |
| NFATC4 | TCEAL6 | 0.767112 | 3 | 0 | 3 |
| NFATC4 | C19orf47 | 0.755493 | 3 | 0 | 3 |
| NFATC4 | CYP21A2 | 0.744848 | 3 | 0 | 3 |
| NFATC4 | GPRASP2 | 0.742446 | 3 | 0 | 3 |
| NFATC4 | ZEB1 | 0.741034 | 3 | 0 | 3 |
| NFATC4 | AP1S2 | 0.740454 | 3 | 0 | 3 |
| NFATC4 | SLC39A3 | 0.738178 | 3 | 0 | 3 |
| NFATC4 | NR2F1 | 0.734909 | 3 | 0 | 3 |
| NFATC4 | RBM38 | 0.72812 | 3 | 0 | 3 |
| NFATC4 | KLHL30 | 0.728018 | 3 | 0 | 3 |
| NFATC4 | EID1 | 0.725543 | 4 | 0 | 4 |
| NFATC4 | RHOJ | 0.721737 | 4 | 0 | 3 |
| NFATC4 | FNDC5 | 0.715024 | 3 | 0 | 3 |
| NFATC4 | CTSO | 0.713401 | 4 | 0 | 4 |
| NFATC4 | PARD3B | 0.712937 | 5 | 0 | 5 |
| NFATC4 | PIEZO1 | 0.711247 | 3 | 0 | 3 |
For details and further investigation, click here