| Full name: ninein | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 14q22.1 | ||
| Entrez ID: 51199 | HGNC ID: HGNC:14906 | Ensembl Gene: ENSG00000100503 | OMIM ID: 608684 |
| Drug and gene relationship at DGIdb | |||
Expression of NIN:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NIN | 51199 | 225921_at | 0.4071 | 0.3622 | |
| GSE20347 | NIN | 51199 | 219285_s_at | -0.1045 | 0.1421 | |
| GSE23400 | NIN | 51199 | 219285_s_at | -0.0820 | 0.0045 | |
| GSE26886 | NIN | 51199 | 225921_at | 0.5631 | 0.0637 | |
| GSE29001 | NIN | 51199 | 219285_s_at | -0.2945 | 0.0421 | |
| GSE38129 | NIN | 51199 | 219285_s_at | -0.1361 | 0.0198 | |
| GSE45670 | NIN | 51199 | 225921_at | 0.0427 | 0.8636 | |
| GSE53622 | NIN | 51199 | 12471 | 0.3048 | 0.0000 | |
| GSE53624 | NIN | 51199 | 16166 | 0.2596 | 0.0001 | |
| GSE63941 | NIN | 51199 | 225921_at | -1.0621 | 0.0303 | |
| GSE77861 | NIN | 51199 | 225921_at | 0.7193 | 0.0687 | |
| GSE97050 | NIN | 51199 | A_24_P412512 | 0.4965 | 0.1557 | |
| SRP007169 | NIN | 51199 | RNAseq | 1.7161 | 0.0000 | |
| SRP008496 | NIN | 51199 | RNAseq | 1.4484 | 0.0000 | |
| SRP064894 | NIN | 51199 | RNAseq | 0.3493 | 0.0904 | |
| SRP133303 | NIN | 51199 | RNAseq | 0.1315 | 0.2545 | |
| SRP159526 | NIN | 51199 | RNAseq | -0.0288 | 0.9149 | |
| SRP193095 | NIN | 51199 | RNAseq | 0.2760 | 0.0934 | |
| SRP219564 | NIN | 51199 | RNAseq | 0.2975 | 0.4625 | |
| TCGA | NIN | 51199 | RNAseq | -0.0240 | 0.6302 |
Upregulated datasets: 2; Downregulated datasets: 1.
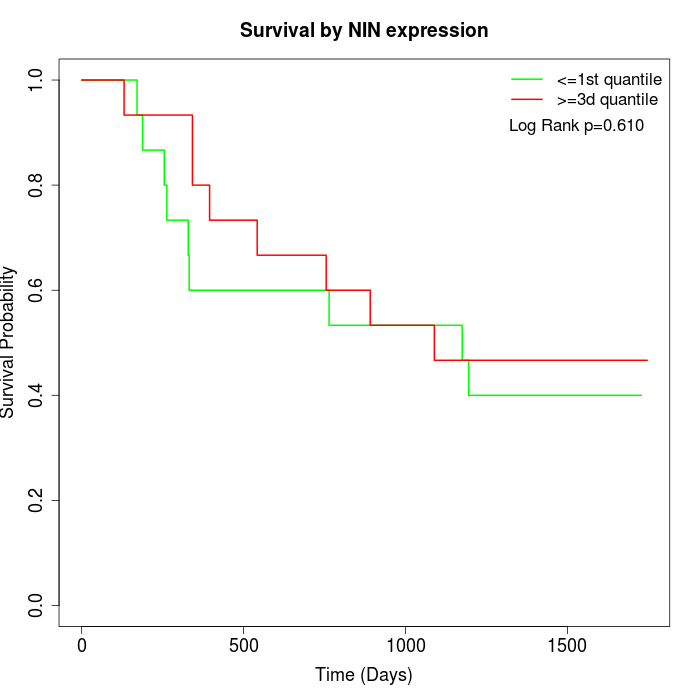
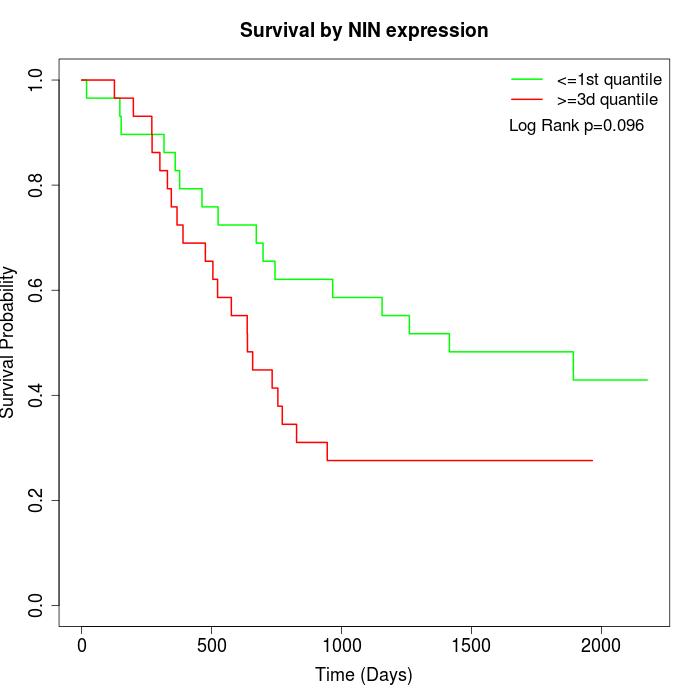
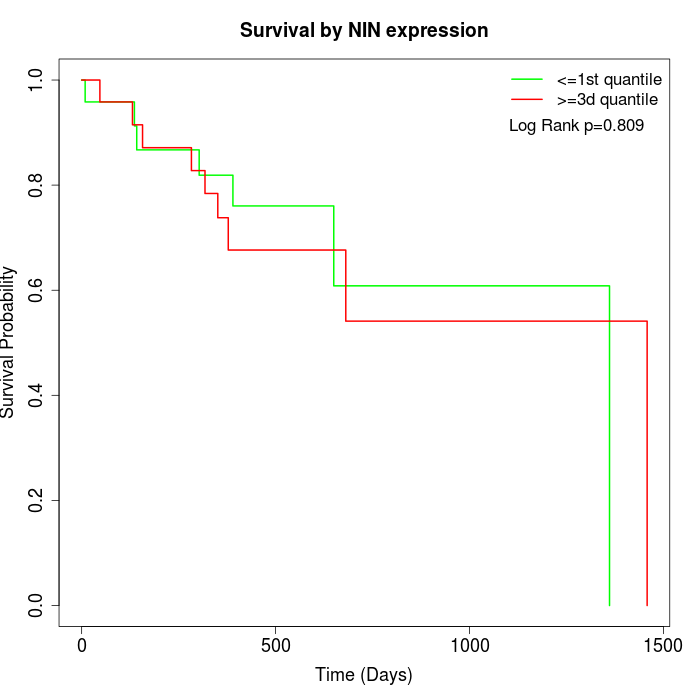
Survival by NIN expression:
Note: Click image to view full size file.
Copy number change of NIN:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NIN | 51199 | 8 | 2 | 20 | |
| GSE20123 | NIN | 51199 | 8 | 3 | 19 | |
| GSE43470 | NIN | 51199 | 9 | 1 | 33 | |
| GSE46452 | NIN | 51199 | 16 | 3 | 40 | |
| GSE47630 | NIN | 51199 | 11 | 10 | 19 | |
| GSE54993 | NIN | 51199 | 3 | 9 | 58 | |
| GSE54994 | NIN | 51199 | 19 | 4 | 30 | |
| GSE60625 | NIN | 51199 | 0 | 2 | 9 | |
| GSE74703 | NIN | 51199 | 8 | 1 | 27 | |
| GSE74704 | NIN | 51199 | 3 | 2 | 15 | |
| TCGA | NIN | 51199 | 34 | 13 | 49 |
Total number of gains: 119; Total number of losses: 50; Total Number of normals: 319.
Somatic mutations of NIN:
Generating mutation plots.
Highly correlated genes for NIN:
Showing top 20/325 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NIN | SMCR8 | 0.800379 | 4 | 0 | 4 |
| NIN | ZNF324B | 0.788724 | 3 | 0 | 3 |
| NIN | PWWP2A | 0.77714 | 3 | 0 | 3 |
| NIN | NPHP3 | 0.76604 | 3 | 0 | 3 |
| NIN | NEMF | 0.738754 | 3 | 0 | 3 |
| NIN | PTAR1 | 0.736169 | 3 | 0 | 3 |
| NIN | TDG | 0.728786 | 3 | 0 | 3 |
| NIN | TRIM35 | 0.726646 | 3 | 0 | 3 |
| NIN | ACBD5 | 0.725508 | 3 | 0 | 3 |
| NIN | CALCRL | 0.71038 | 3 | 0 | 3 |
| NIN | CBFA2T2 | 0.709356 | 3 | 0 | 3 |
| NIN | TMTC1 | 0.707054 | 4 | 0 | 4 |
| NIN | MAPKAPK2 | 0.699443 | 3 | 0 | 3 |
| NIN | PTPMT1 | 0.697534 | 4 | 0 | 4 |
| NIN | TPM3 | 0.694056 | 3 | 0 | 3 |
| NIN | TMTC3 | 0.692654 | 5 | 0 | 5 |
| NIN | TRUB1 | 0.68836 | 5 | 0 | 4 |
| NIN | C12orf57 | 0.688052 | 4 | 0 | 3 |
| NIN | VMA21 | 0.685528 | 5 | 0 | 4 |
| NIN | RHBDD2 | 0.684116 | 3 | 0 | 3 |
For details and further investigation, click here