| Full name: NRAS proto-oncogene, GTPase | Alias Symbol: N-ras | ||
| Type: protein-coding gene | Cytoband: 1p13.2 | ||
| Entrez ID: 4893 | HGNC ID: HGNC:7989 | Ensembl Gene: ENSG00000213281 | OMIM ID: 164790 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
NRAS involved pathways:
Expression of NRAS:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NRAS | 4893 | 224985_at | 0.1195 | 0.8823 | |
| GSE20347 | NRAS | 4893 | 202647_s_at | 0.1422 | 0.6073 | |
| GSE23400 | NRAS | 4893 | 202647_s_at | 0.4664 | 0.0000 | |
| GSE26886 | NRAS | 4893 | 224985_at | -0.3537 | 0.0304 | |
| GSE29001 | NRAS | 4893 | 202647_s_at | 0.1317 | 0.7436 | |
| GSE38129 | NRAS | 4893 | 202647_s_at | 0.5086 | 0.0155 | |
| GSE45670 | NRAS | 4893 | 224985_at | 0.2529 | 0.0497 | |
| GSE53622 | NRAS | 4893 | 10919 | 0.4486 | 0.0000 | |
| GSE53624 | NRAS | 4893 | 10919 | 0.3755 | 0.0000 | |
| GSE63941 | NRAS | 4893 | 224985_at | 0.1194 | 0.8572 | |
| GSE77861 | NRAS | 4893 | 224985_at | -0.2591 | 0.1964 | |
| GSE97050 | NRAS | 4893 | A_33_P3317825 | 0.6427 | 0.2204 | |
| SRP007169 | NRAS | 4893 | RNAseq | -0.2435 | 0.5060 | |
| SRP008496 | NRAS | 4893 | RNAseq | -0.2307 | 0.2579 | |
| SRP064894 | NRAS | 4893 | RNAseq | 0.1154 | 0.5856 | |
| SRP133303 | NRAS | 4893 | RNAseq | 0.5090 | 0.0085 | |
| SRP159526 | NRAS | 4893 | RNAseq | -0.5229 | 0.0307 | |
| SRP193095 | NRAS | 4893 | RNAseq | -0.1196 | 0.3770 | |
| SRP219564 | NRAS | 4893 | RNAseq | -0.0415 | 0.9460 | |
| TCGA | NRAS | 4893 | RNAseq | 0.1475 | 0.0049 |
Upregulated datasets: 0; Downregulated datasets: 0.
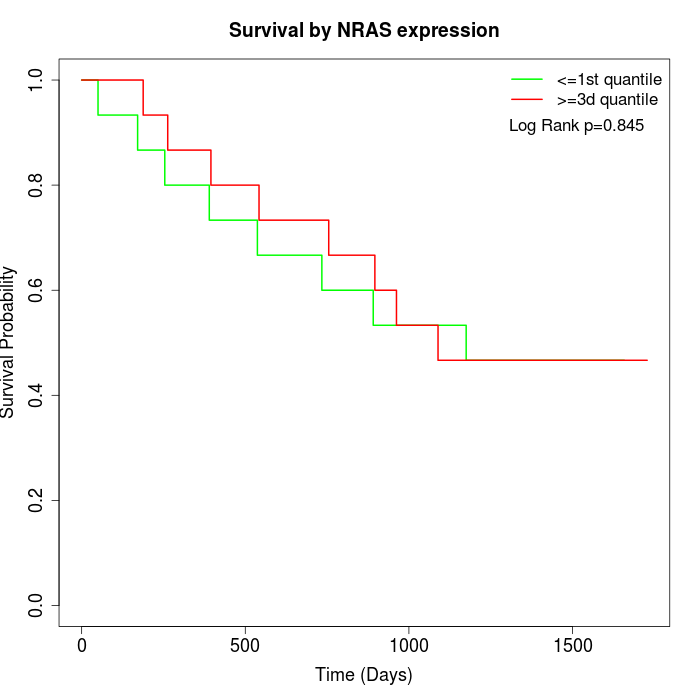
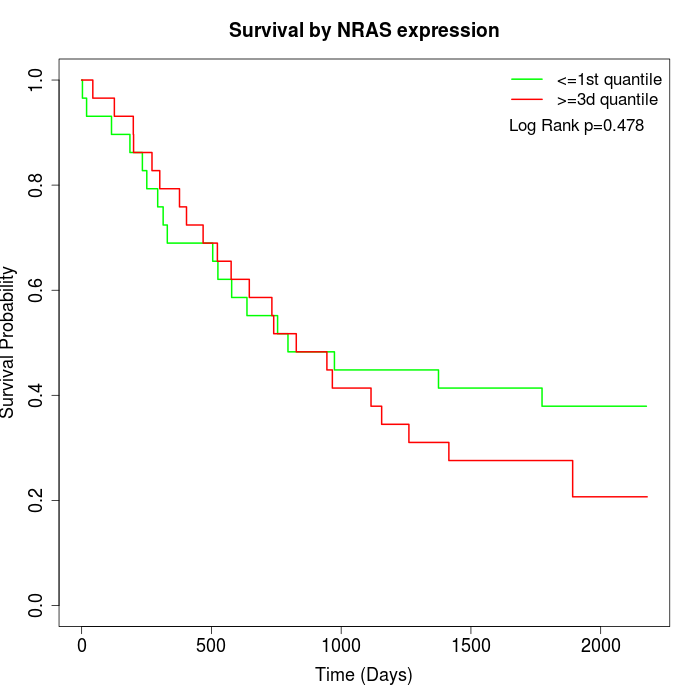
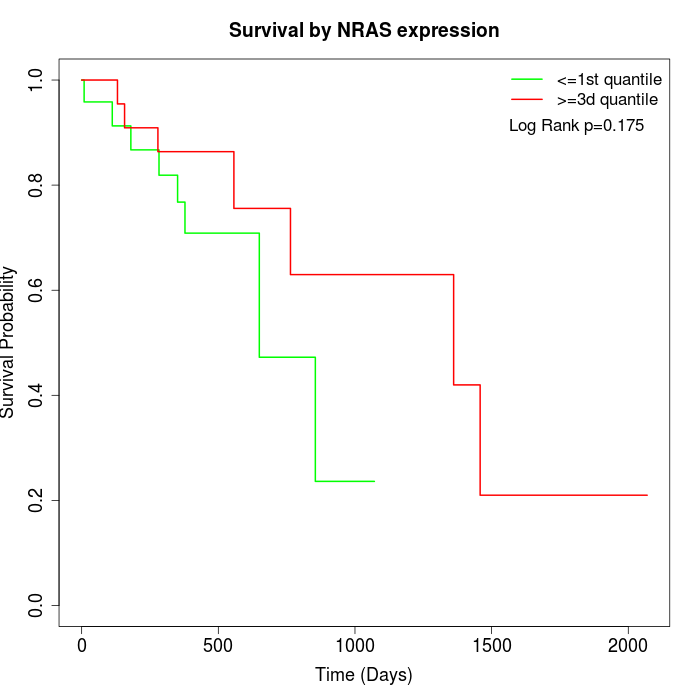
Survival by NRAS expression:
Note: Click image to view full size file.
Copy number change of NRAS:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NRAS | 4893 | 0 | 9 | 21 | |
| GSE20123 | NRAS | 4893 | 0 | 9 | 21 | |
| GSE43470 | NRAS | 4893 | 0 | 8 | 35 | |
| GSE46452 | NRAS | 4893 | 2 | 1 | 56 | |
| GSE47630 | NRAS | 4893 | 9 | 5 | 26 | |
| GSE54993 | NRAS | 4893 | 0 | 1 | 69 | |
| GSE54994 | NRAS | 4893 | 7 | 3 | 43 | |
| GSE60625 | NRAS | 4893 | 0 | 0 | 11 | |
| GSE74703 | NRAS | 4893 | 0 | 7 | 29 | |
| GSE74704 | NRAS | 4893 | 0 | 5 | 15 | |
| TCGA | NRAS | 4893 | 12 | 30 | 54 |
Total number of gains: 30; Total number of losses: 78; Total Number of normals: 380.
Somatic mutations of NRAS:
Generating mutation plots.
Highly correlated genes for NRAS:
Showing top 20/434 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NRAS | TRIB3 | 0.735328 | 4 | 0 | 4 |
| NRAS | TMEM54 | 0.715016 | 3 | 0 | 3 |
| NRAS | GALE | 0.697015 | 3 | 0 | 3 |
| NRAS | FAM83B | 0.689746 | 5 | 0 | 5 |
| NRAS | AMPD3 | 0.669635 | 4 | 0 | 4 |
| NRAS | SPAG1 | 0.658007 | 5 | 0 | 4 |
| NRAS | GSTO2 | 0.655186 | 3 | 0 | 3 |
| NRAS | AJUBA | 0.654998 | 4 | 0 | 3 |
| NRAS | WNT10A | 0.654841 | 4 | 0 | 3 |
| NRAS | ETV4 | 0.650786 | 5 | 0 | 3 |
| NRAS | MYSM1 | 0.647144 | 3 | 0 | 3 |
| NRAS | SPHK1 | 0.640589 | 5 | 0 | 4 |
| NRAS | IL1RAP | 0.632524 | 9 | 0 | 7 |
| NRAS | EPHA2 | 0.632328 | 4 | 0 | 3 |
| NRAS | VAV3 | 0.631615 | 3 | 0 | 3 |
| NRAS | GJB3 | 0.631406 | 6 | 0 | 4 |
| NRAS | IRAK2 | 0.628227 | 4 | 0 | 3 |
| NRAS | CNIH1 | 0.628007 | 5 | 0 | 4 |
| NRAS | OLFM2 | 0.62584 | 3 | 0 | 3 |
| NRAS | SRP9 | 0.62443 | 3 | 0 | 3 |
For details and further investigation, click here