| Full name: EPH receptor A2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 1p36.13 | ||
| Entrez ID: 1969 | HGNC ID: HGNC:3386 | Ensembl Gene: ENSG00000142627 | OMIM ID: 176946 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
EPHA2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04014 | Ras signaling pathway | |
| hsa04015 | Rap1 signaling pathway | |
| hsa04151 | PI3K-Akt signaling pathway | |
| hsa04360 | Axon guidance |
Expression of EPHA2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | EPHA2 | 1969 | 203499_at | -0.5987 | 0.5794 | |
| GSE20347 | EPHA2 | 1969 | 203499_at | -1.2258 | 0.0000 | |
| GSE23400 | EPHA2 | 1969 | 203499_at | -1.2130 | 0.0000 | |
| GSE26886 | EPHA2 | 1969 | 203499_at | -2.4339 | 0.0000 | |
| GSE29001 | EPHA2 | 1969 | 203499_at | -1.2986 | 0.0004 | |
| GSE38129 | EPHA2 | 1969 | 203499_at | -0.6739 | 0.0253 | |
| GSE45670 | EPHA2 | 1969 | 203499_at | -0.0136 | 0.9687 | |
| GSE53622 | EPHA2 | 1969 | 3426 | -1.1688 | 0.0000 | |
| GSE53624 | EPHA2 | 1969 | 3426 | -1.6189 | 0.0000 | |
| GSE63941 | EPHA2 | 1969 | 203499_at | 1.1779 | 0.0592 | |
| GSE77861 | EPHA2 | 1969 | 203499_at | -0.9423 | 0.0018 | |
| GSE97050 | EPHA2 | 1969 | A_23_P149281 | 0.4814 | 0.5688 | |
| SRP007169 | EPHA2 | 1969 | RNAseq | -3.1365 | 0.0000 | |
| SRP008496 | EPHA2 | 1969 | RNAseq | -3.3758 | 0.0000 | |
| SRP064894 | EPHA2 | 1969 | RNAseq | -2.1896 | 0.0000 | |
| SRP133303 | EPHA2 | 1969 | RNAseq | -1.2508 | 0.0000 | |
| SRP159526 | EPHA2 | 1969 | RNAseq | -2.1683 | 0.0000 | |
| SRP193095 | EPHA2 | 1969 | RNAseq | -1.4158 | 0.0000 | |
| SRP219564 | EPHA2 | 1969 | RNAseq | -2.2302 | 0.0022 | |
| TCGA | EPHA2 | 1969 | RNAseq | 0.2080 | 0.0344 |
Upregulated datasets: 0; Downregulated datasets: 13.
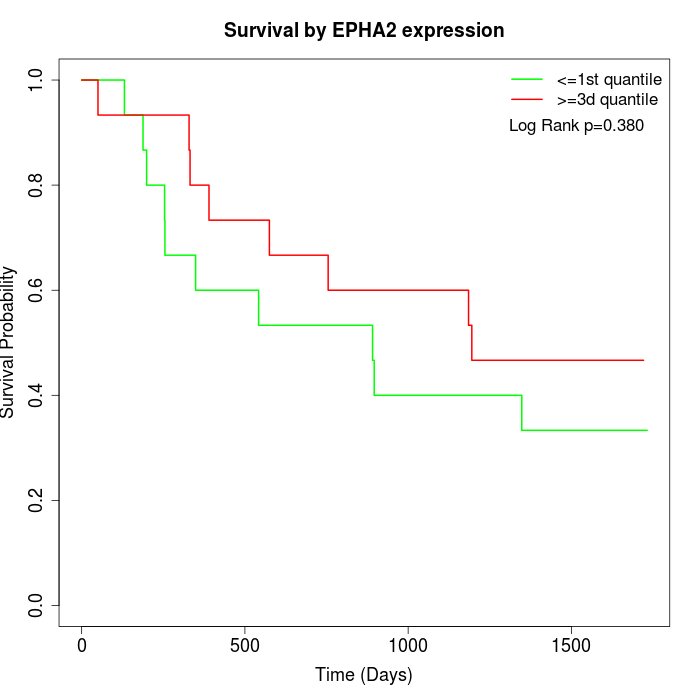
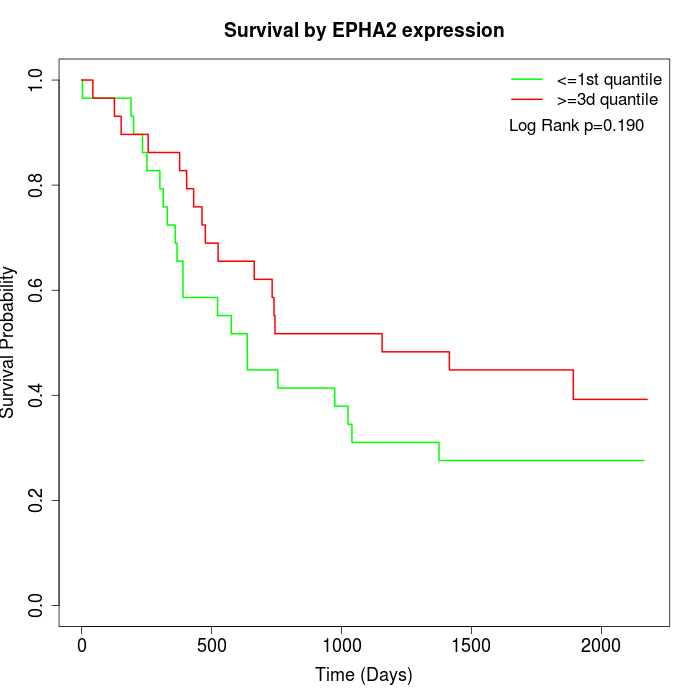
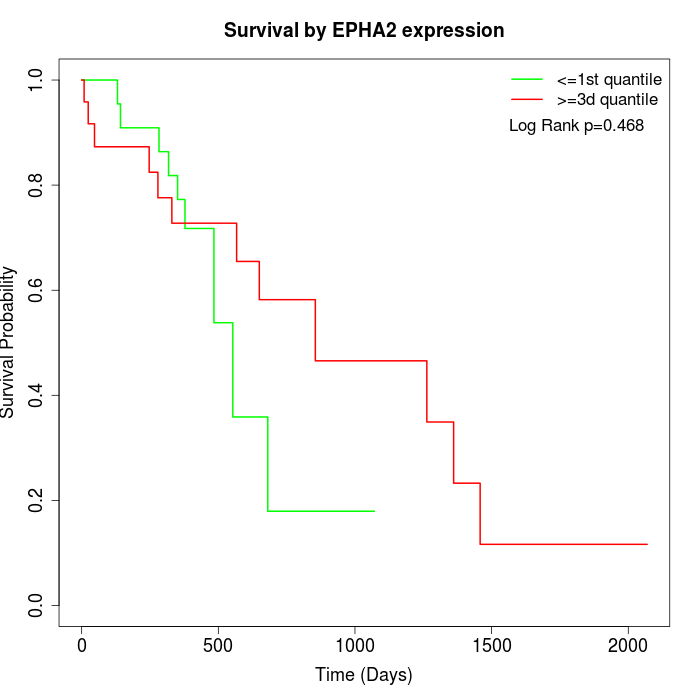
Survival by EPHA2 expression:
Note: Click image to view full size file.
Copy number change of EPHA2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | EPHA2 | 1969 | 1 | 5 | 24 | |
| GSE20123 | EPHA2 | 1969 | 1 | 4 | 25 | |
| GSE43470 | EPHA2 | 1969 | 1 | 8 | 34 | |
| GSE46452 | EPHA2 | 1969 | 7 | 1 | 51 | |
| GSE47630 | EPHA2 | 1969 | 8 | 3 | 29 | |
| GSE54993 | EPHA2 | 1969 | 3 | 1 | 66 | |
| GSE54994 | EPHA2 | 1969 | 10 | 5 | 38 | |
| GSE60625 | EPHA2 | 1969 | 0 | 0 | 11 | |
| GSE74703 | EPHA2 | 1969 | 1 | 5 | 30 | |
| GSE74704 | EPHA2 | 1969 | 1 | 0 | 19 | |
| TCGA | EPHA2 | 1969 | 11 | 24 | 61 |
Total number of gains: 44; Total number of losses: 56; Total Number of normals: 388.
Somatic mutations of EPHA2:
Generating mutation plots.
Highly correlated genes for EPHA2:
Showing top 20/1436 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| EPHA2 | NUAK2 | 0.817342 | 4 | 0 | 4 |
| EPHA2 | NIPAL1 | 0.80489 | 3 | 0 | 3 |
| EPHA2 | TGM1 | 0.785592 | 12 | 0 | 12 |
| EPHA2 | MXD1 | 0.777973 | 11 | 0 | 11 |
| EPHA2 | CYSRT1 | 0.774645 | 6 | 0 | 6 |
| EPHA2 | PLIN3 | 0.772393 | 11 | 0 | 11 |
| EPHA2 | EPS8L2 | 0.766091 | 10 | 0 | 9 |
| EPHA2 | IL1RN | 0.765586 | 12 | 0 | 11 |
| EPHA2 | PTK6 | 0.761001 | 11 | 0 | 10 |
| EPHA2 | DUOX1 | 0.760973 | 12 | 0 | 12 |
| EPHA2 | SPRR2F | 0.760083 | 3 | 0 | 3 |
| EPHA2 | CRCT1 | 0.75918 | 11 | 0 | 11 |
| EPHA2 | SH3GL1 | 0.75862 | 11 | 0 | 11 |
| EPHA2 | RAB10 | 0.757155 | 6 | 0 | 6 |
| EPHA2 | DUSP5 | 0.756254 | 13 | 0 | 11 |
| EPHA2 | RAET1E | 0.756205 | 6 | 0 | 6 |
| EPHA2 | SULT2B1 | 0.754652 | 9 | 0 | 9 |
| EPHA2 | PRSS22 | 0.752986 | 12 | 0 | 12 |
| EPHA2 | IL36A | 0.752126 | 10 | 0 | 9 |
| EPHA2 | DENND2C | 0.750586 | 8 | 0 | 8 |
For details and further investigation, click here