| Full name: neuregulin 3 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 10q23.1 | ||
| Entrez ID: 10718 | HGNC ID: HGNC:7999 | Ensembl Gene: ENSG00000185737 | OMIM ID: 605533 |
| Drug and gene relationship at DGIdb | |||
NRG3 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04012 | ErbB signaling pathway |
Expression of NRG3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NRG3 | 10718 | 229233_at | -0.0346 | 0.9194 | |
| GSE26886 | NRG3 | 10718 | 229233_at | 0.0817 | 0.3569 | |
| GSE45670 | NRG3 | 10718 | 229233_at | -0.1938 | 0.0217 | |
| GSE53622 | NRG3 | 10718 | 8213 | -0.2116 | 0.1058 | |
| GSE53624 | NRG3 | 10718 | 8213 | -0.0070 | 0.9496 | |
| GSE63941 | NRG3 | 10718 | 229233_at | 0.0037 | 0.9839 | |
| GSE77861 | NRG3 | 10718 | 229233_at | -0.0762 | 0.4298 | |
| GSE97050 | NRG3 | 10718 | A_33_P3229417 | 0.0714 | 0.7673 | |
| SRP133303 | NRG3 | 10718 | RNAseq | -1.4766 | 0.0001 | |
| TCGA | NRG3 | 10718 | RNAseq | -2.6507 | 0.0031 |
Upregulated datasets: 0; Downregulated datasets: 2.
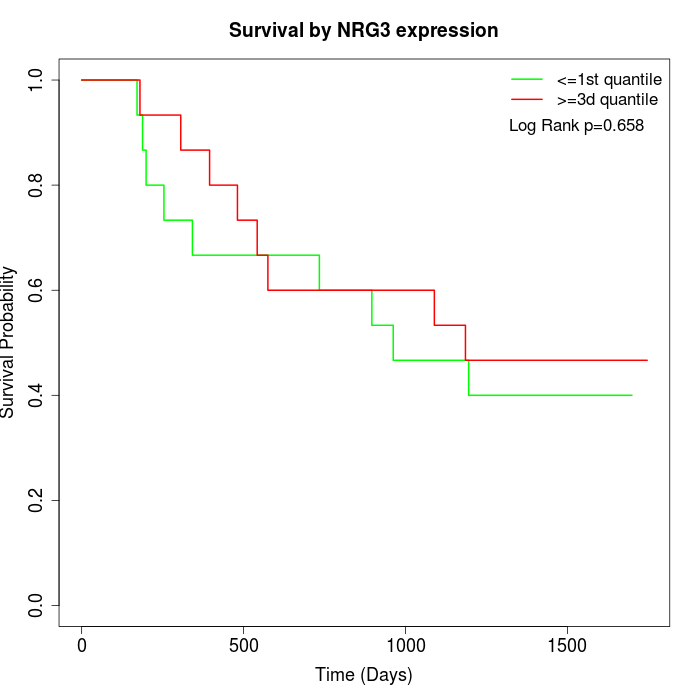
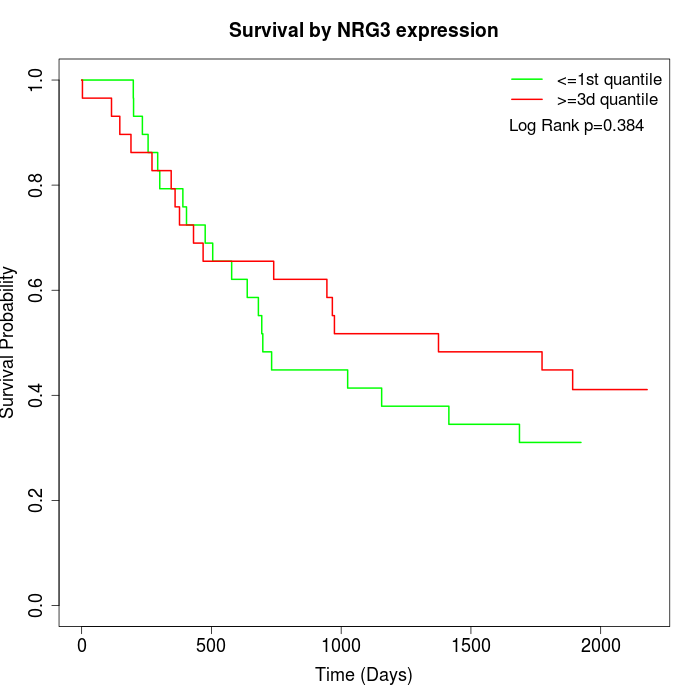
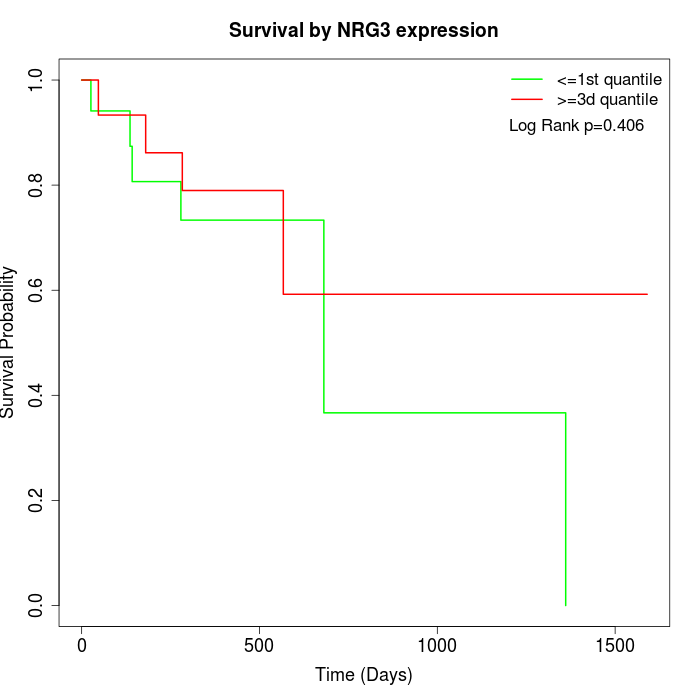
Survival by NRG3 expression:
Note: Click image to view full size file.
Copy number change of NRG3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NRG3 | 10718 | 2 | 6 | 22 | |
| GSE20123 | NRG3 | 10718 | 2 | 5 | 23 | |
| GSE43470 | NRG3 | 10718 | 1 | 6 | 36 | |
| GSE46452 | NRG3 | 10718 | 0 | 11 | 48 | |
| GSE47630 | NRG3 | 10718 | 2 | 14 | 24 | |
| GSE54993 | NRG3 | 10718 | 7 | 0 | 63 | |
| GSE54994 | NRG3 | 10718 | 2 | 11 | 40 | |
| GSE60625 | NRG3 | 10718 | 0 | 0 | 11 | |
| GSE74703 | NRG3 | 10718 | 1 | 4 | 31 | |
| GSE74704 | NRG3 | 10718 | 1 | 3 | 16 | |
| TCGA | NRG3 | 10718 | 10 | 24 | 62 |
Total number of gains: 28; Total number of losses: 84; Total Number of normals: 376.
Somatic mutations of NRG3:
Generating mutation plots.
Highly correlated genes for NRG3:
Showing top 20/131 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NRG3 | COL20A1 | 0.767879 | 3 | 0 | 3 |
| NRG3 | TULP2 | 0.728276 | 3 | 0 | 3 |
| NRG3 | RPH3A | 0.726163 | 3 | 0 | 3 |
| NRG3 | KIF17 | 0.719731 | 3 | 0 | 3 |
| NRG3 | TRPV3 | 0.709063 | 4 | 0 | 4 |
| NRG3 | SIX3 | 0.704288 | 3 | 0 | 3 |
| NRG3 | UROC1 | 0.70028 | 3 | 0 | 3 |
| NRG3 | CRYGA | 0.681561 | 3 | 0 | 3 |
| NRG3 | AMHR2 | 0.67848 | 3 | 0 | 3 |
| NRG3 | CLDN9 | 0.67448 | 4 | 0 | 4 |
| NRG3 | CSHL1 | 0.669564 | 3 | 0 | 3 |
| NRG3 | HDAC10 | 0.66611 | 3 | 0 | 3 |
| NRG3 | MIXL1 | 0.665656 | 3 | 0 | 3 |
| NRG3 | RGL4 | 0.660418 | 4 | 0 | 3 |
| NRG3 | TRIM10 | 0.658727 | 4 | 0 | 3 |
| NRG3 | FAM131A | 0.657429 | 3 | 0 | 3 |
| NRG3 | NXNL1 | 0.65458 | 4 | 0 | 3 |
| NRG3 | TSSK3 | 0.652861 | 4 | 0 | 4 |
| NRG3 | TRH | 0.650518 | 4 | 0 | 3 |
| NRG3 | ADIG | 0.647303 | 3 | 0 | 3 |
For details and further investigation, click here