| Full name: phosphate cytidylyltransferase 1, choline, beta | Alias Symbol: CCT-beta|CTB | ||
| Type: protein-coding gene | Cytoband: Xp22.11 | ||
| Entrez ID: 9468 | HGNC ID: HGNC:8755 | Ensembl Gene: ENSG00000102230 | OMIM ID: 300948 |
| Drug and gene relationship at DGIdb | |||
PCYT1B involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05231 | Choline metabolism in cancer |
Expression of PCYT1B:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PCYT1B | 9468 | 210456_at | 0.0003 | 0.9994 | |
| GSE20347 | PCYT1B | 9468 | 210456_at | 0.1515 | 0.0628 | |
| GSE23400 | PCYT1B | 9468 | 210456_at | -0.0320 | 0.1375 | |
| GSE26886 | PCYT1B | 9468 | 210456_at | 0.2666 | 0.0250 | |
| GSE29001 | PCYT1B | 9468 | 206751_s_at | 0.2556 | 0.2291 | |
| GSE38129 | PCYT1B | 9468 | 210456_at | 0.0747 | 0.3076 | |
| GSE45670 | PCYT1B | 9468 | 232553_at | 0.0014 | 0.9948 | |
| GSE53622 | PCYT1B | 9468 | 20833 | 0.1867 | 0.4304 | |
| GSE53624 | PCYT1B | 9468 | 57942 | 0.3337 | 0.1310 | |
| GSE63941 | PCYT1B | 9468 | 210456_at | 0.0243 | 0.9047 | |
| GSE77861 | PCYT1B | 9468 | 210456_at | 0.0037 | 0.9880 | |
| GSE97050 | PCYT1B | 9468 | A_23_P148422 | -0.1728 | 0.6443 | |
| SRP007169 | PCYT1B | 9468 | RNAseq | 2.0567 | 0.0149 | |
| SRP064894 | PCYT1B | 9468 | RNAseq | -0.1070 | 0.6937 | |
| SRP133303 | PCYT1B | 9468 | RNAseq | 0.6089 | 0.1690 | |
| SRP159526 | PCYT1B | 9468 | RNAseq | 1.1250 | 0.0130 | |
| SRP193095 | PCYT1B | 9468 | RNAseq | 1.2490 | 0.0000 | |
| SRP219564 | PCYT1B | 9468 | RNAseq | -0.4070 | 0.4929 | |
| TCGA | PCYT1B | 9468 | RNAseq | 0.6556 | 0.0467 |
Upregulated datasets: 3; Downregulated datasets: 0.
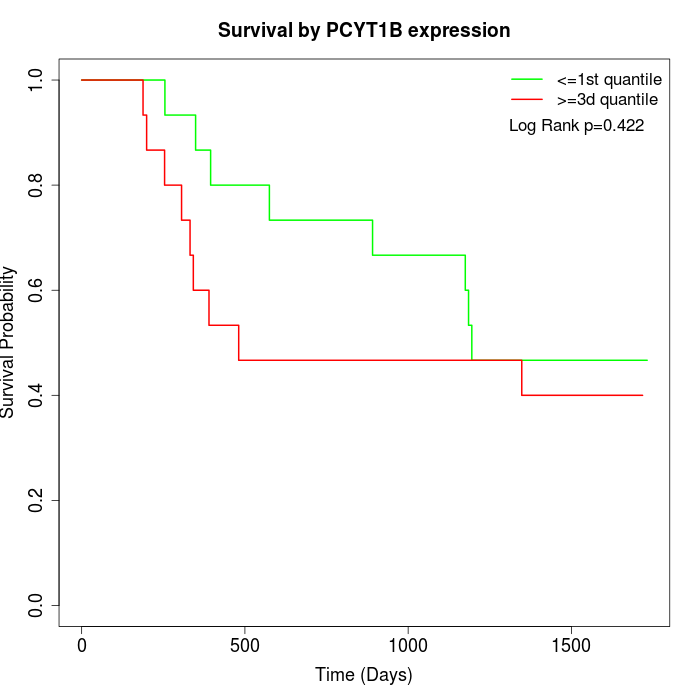
Survival by PCYT1B expression:
Note: Click image to view full size file.
Copy number change of PCYT1B:
No record found for this gene.
Somatic mutations of PCYT1B:
Generating mutation plots.
Highly correlated genes for PCYT1B:
Showing top 20/148 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PCYT1B | TCAP | 0.730513 | 3 | 0 | 3 |
| PCYT1B | NFAM1 | 0.710274 | 3 | 0 | 3 |
| PCYT1B | CALN1 | 0.701837 | 3 | 0 | 3 |
| PCYT1B | ERF | 0.692644 | 3 | 0 | 3 |
| PCYT1B | PAGE5 | 0.683936 | 3 | 0 | 3 |
| PCYT1B | FAM167B | 0.67516 | 3 | 0 | 3 |
| PCYT1B | DERL3 | 0.671881 | 4 | 0 | 4 |
| PCYT1B | HMCN2 | 0.670872 | 3 | 0 | 3 |
| PCYT1B | ADAMTSL5 | 0.667299 | 3 | 0 | 3 |
| PCYT1B | FGF12 | 0.66714 | 3 | 0 | 3 |
| PCYT1B | CCDC85A | 0.663557 | 3 | 0 | 3 |
| PCYT1B | ZDHHC1 | 0.659395 | 4 | 0 | 4 |
| PCYT1B | TOX | 0.658723 | 4 | 0 | 3 |
| PCYT1B | CCDC169 | 0.641697 | 4 | 0 | 4 |
| PCYT1B | LDB3 | 0.638608 | 3 | 0 | 3 |
| PCYT1B | FIBCD1 | 0.632503 | 3 | 0 | 3 |
| PCYT1B | ZNF579 | 0.624173 | 3 | 0 | 3 |
| PCYT1B | CCDC78 | 0.623476 | 4 | 0 | 4 |
| PCYT1B | EGFLAM | 0.622477 | 3 | 0 | 3 |
| PCYT1B | C9orf50 | 0.621875 | 4 | 0 | 3 |
For details and further investigation, click here