| Full name: thymocyte selection associated high mobility group box | Alias Symbol: KIAA0808|TOX1 | ||
| Type: protein-coding gene | Cytoband: 8q12.1 | ||
| Entrez ID: 9760 | HGNC ID: HGNC:18988 | Ensembl Gene: ENSG00000198846 | OMIM ID: 606863 |
| Drug and gene relationship at DGIdb | |||
Expression of TOX:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TOX | 9760 | 204529_s_at | -0.8512 | 0.5912 | |
| GSE20347 | TOX | 9760 | 204529_s_at | 0.2673 | 0.3742 | |
| GSE23400 | TOX | 9760 | 204529_s_at | -0.1143 | 0.2521 | |
| GSE26886 | TOX | 9760 | 204529_s_at | 1.4622 | 0.0026 | |
| GSE29001 | TOX | 9760 | 204529_s_at | 0.3561 | 0.1662 | |
| GSE38129 | TOX | 9760 | 204529_s_at | -0.3936 | 0.4019 | |
| GSE45670 | TOX | 9760 | 204529_s_at | -1.1711 | 0.0263 | |
| GSE53622 | TOX | 9760 | 21917 | -1.2340 | 0.0000 | |
| GSE53624 | TOX | 9760 | 21917 | -0.9803 | 0.0000 | |
| GSE63941 | TOX | 9760 | 204529_s_at | -1.6145 | 0.3847 | |
| GSE77861 | TOX | 9760 | 204530_s_at | -0.1144 | 0.2860 | |
| GSE97050 | TOX | 9760 | A_24_P226755 | -0.4387 | 0.3269 | |
| SRP007169 | TOX | 9760 | RNAseq | 3.6763 | 0.0001 | |
| SRP064894 | TOX | 9760 | RNAseq | -0.4555 | 0.1177 | |
| SRP133303 | TOX | 9760 | RNAseq | -0.2508 | 0.5418 | |
| SRP159526 | TOX | 9760 | RNAseq | -0.5916 | 0.1967 | |
| SRP219564 | TOX | 9760 | RNAseq | 0.2481 | 0.7900 | |
| TCGA | TOX | 9760 | RNAseq | -0.9632 | 0.0000 |
Upregulated datasets: 2; Downregulated datasets: 2.
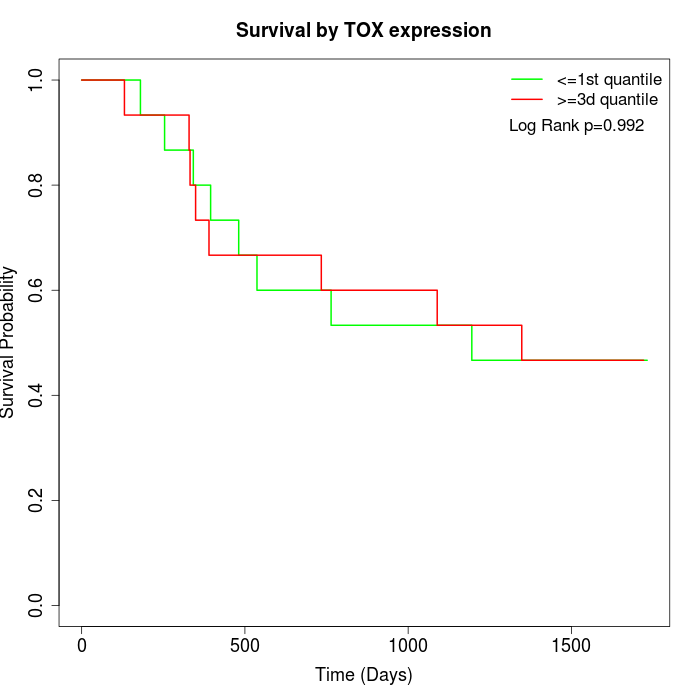
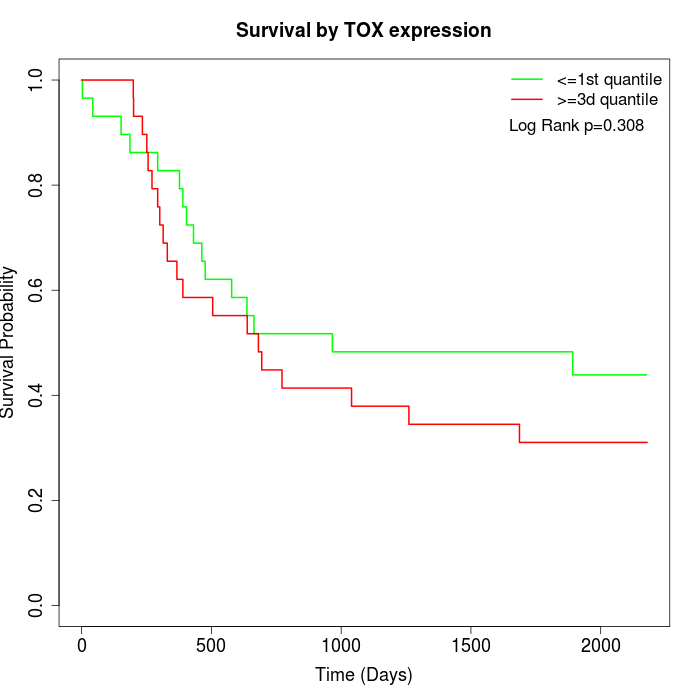
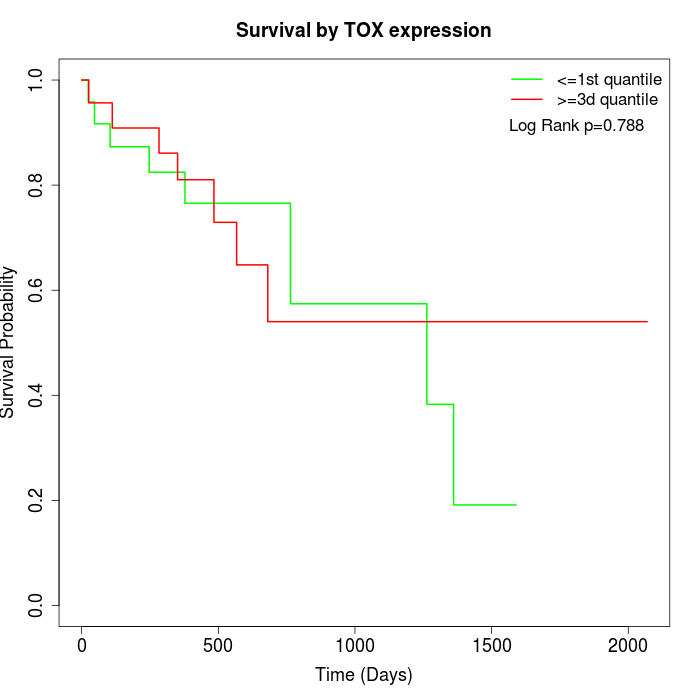
Survival by TOX expression:
Note: Click image to view full size file.
Copy number change of TOX:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TOX | 9760 | 13 | 1 | 16 | |
| GSE20123 | TOX | 9760 | 13 | 1 | 16 | |
| GSE43470 | TOX | 9760 | 14 | 3 | 26 | |
| GSE46452 | TOX | 9760 | 20 | 3 | 36 | |
| GSE47630 | TOX | 9760 | 23 | 1 | 16 | |
| GSE54993 | TOX | 9760 | 1 | 17 | 52 | |
| GSE54994 | TOX | 9760 | 30 | 0 | 23 | |
| GSE60625 | TOX | 9760 | 0 | 4 | 7 | |
| GSE74703 | TOX | 9760 | 12 | 2 | 22 | |
| GSE74704 | TOX | 9760 | 9 | 0 | 11 | |
| TCGA | TOX | 9760 | 44 | 6 | 46 |
Total number of gains: 179; Total number of losses: 38; Total Number of normals: 271.
Somatic mutations of TOX:
Generating mutation plots.
Highly correlated genes for TOX:
Showing top 20/824 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TOX | TIMP3 | 0.751006 | 5 | 0 | 5 |
| TOX | GJC2 | 0.749671 | 3 | 0 | 3 |
| TOX | WDR48 | 0.748215 | 3 | 0 | 3 |
| TOX | GIPC3 | 0.744523 | 3 | 0 | 3 |
| TOX | ZBTB26 | 0.737381 | 3 | 0 | 3 |
| TOX | FXYD1 | 0.733391 | 4 | 0 | 4 |
| TOX | CBX7 | 0.728703 | 7 | 0 | 6 |
| TOX | ILDR2 | 0.719604 | 3 | 0 | 3 |
| TOX | ZFPM2 | 0.716913 | 10 | 0 | 9 |
| TOX | DDX42 | 0.716471 | 4 | 0 | 4 |
| TOX | NEGR1 | 0.713823 | 5 | 0 | 5 |
| TOX | FIBIN | 0.712093 | 3 | 0 | 3 |
| TOX | FGL2 | 0.706537 | 8 | 0 | 6 |
| TOX | WSCD2 | 0.703622 | 3 | 0 | 3 |
| TOX | TCEAL6 | 0.702535 | 3 | 0 | 3 |
| TOX | ZNF599 | 0.701787 | 4 | 0 | 4 |
| TOX | KCNB1 | 0.699922 | 4 | 0 | 4 |
| TOX | KCNN3 | 0.694944 | 6 | 0 | 5 |
| TOX | SMAD9 | 0.694645 | 8 | 0 | 7 |
| TOX | MAMDC2 | 0.694425 | 5 | 0 | 5 |
For details and further investigation, click here