| Full name: phosphodiesterase 1B | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 12q13.2 | ||
| Entrez ID: 5153 | HGNC ID: HGNC:8775 | Ensembl Gene: ENSG00000123360 | OMIM ID: 171891 |
| Drug and gene relationship at DGIdb | |||
PDE1B involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04020 | Calcium signaling pathway | |
| hsa04740 | Olfactory transduction | |
| hsa04742 | Taste transduction | |
| hsa04924 | Renin secretion |
Expression of PDE1B:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PDE1B | 5153 | 206444_at | 0.1631 | 0.4584 | |
| GSE20347 | PDE1B | 5153 | 206444_at | -0.0734 | 0.3168 | |
| GSE23400 | PDE1B | 5153 | 206444_at | -0.0290 | 0.3600 | |
| GSE26886 | PDE1B | 5153 | 206444_at | -0.0049 | 0.9739 | |
| GSE29001 | PDE1B | 5153 | 206444_at | -0.2550 | 0.1189 | |
| GSE38129 | PDE1B | 5153 | 206444_at | -0.1398 | 0.1056 | |
| GSE45670 | PDE1B | 5153 | 206444_at | 0.0615 | 0.5132 | |
| GSE53622 | PDE1B | 5153 | 85248 | -0.6655 | 0.0000 | |
| GSE53624 | PDE1B | 5153 | 85248 | -0.4731 | 0.0000 | |
| GSE63941 | PDE1B | 5153 | 206444_at | 0.4126 | 0.0123 | |
| GSE77861 | PDE1B | 5153 | 206444_at | -0.0850 | 0.4270 | |
| GSE97050 | PDE1B | 5153 | A_23_P139585 | 0.1112 | 0.7673 | |
| SRP064894 | PDE1B | 5153 | RNAseq | 0.1257 | 0.7035 | |
| SRP133303 | PDE1B | 5153 | RNAseq | -0.8230 | 0.0000 | |
| SRP159526 | PDE1B | 5153 | RNAseq | -0.8828 | 0.0073 | |
| SRP193095 | PDE1B | 5153 | RNAseq | 0.4538 | 0.0456 | |
| SRP219564 | PDE1B | 5153 | RNAseq | -0.9130 | 0.0840 | |
| TCGA | PDE1B | 5153 | RNAseq | -0.9433 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 0.
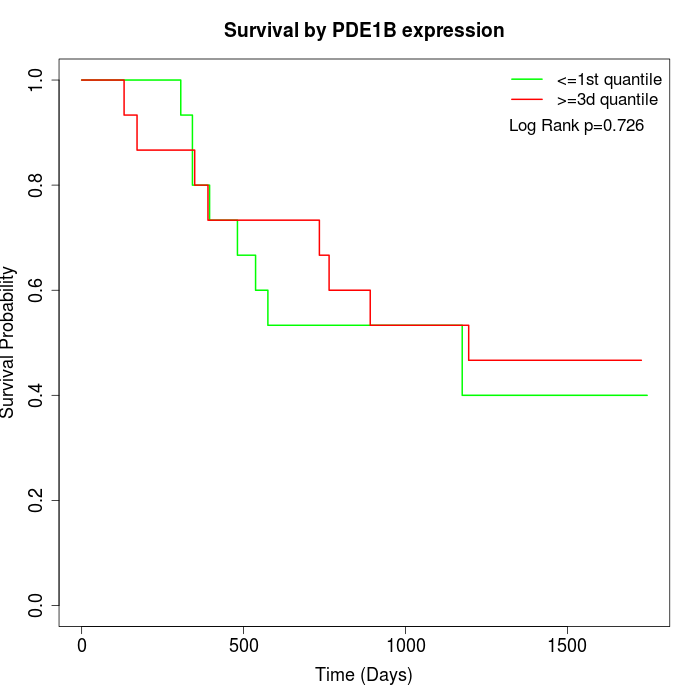
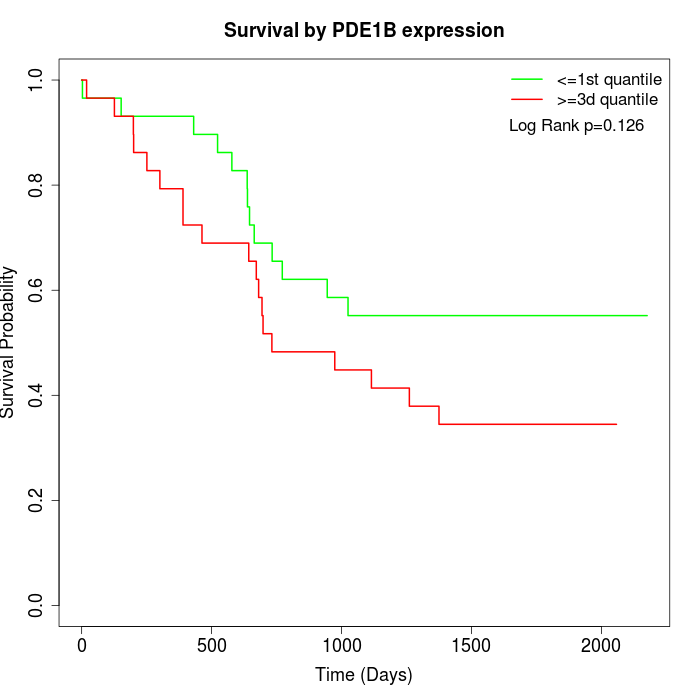
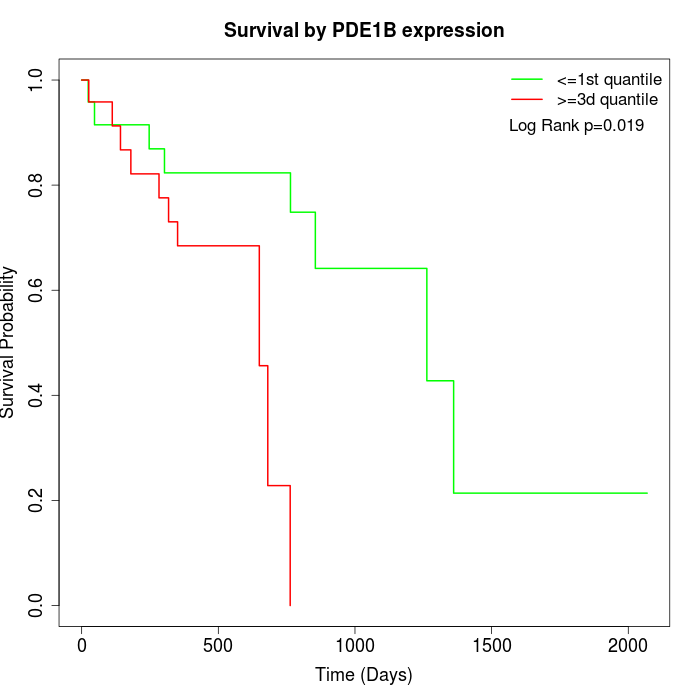
Survival by PDE1B expression:
Note: Click image to view full size file.
Copy number change of PDE1B:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PDE1B | 5153 | 4 | 1 | 25 | |
| GSE20123 | PDE1B | 5153 | 4 | 1 | 25 | |
| GSE43470 | PDE1B | 5153 | 5 | 0 | 38 | |
| GSE46452 | PDE1B | 5153 | 7 | 1 | 51 | |
| GSE47630 | PDE1B | 5153 | 10 | 2 | 28 | |
| GSE54993 | PDE1B | 5153 | 0 | 5 | 65 | |
| GSE54994 | PDE1B | 5153 | 4 | 1 | 48 | |
| GSE60625 | PDE1B | 5153 | 0 | 0 | 11 | |
| GSE74703 | PDE1B | 5153 | 4 | 0 | 32 | |
| GSE74704 | PDE1B | 5153 | 3 | 1 | 16 | |
| TCGA | PDE1B | 5153 | 16 | 11 | 69 |
Total number of gains: 57; Total number of losses: 23; Total Number of normals: 408.
Somatic mutations of PDE1B:
Generating mutation plots.
Highly correlated genes for PDE1B:
Showing top 20/773 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PDE1B | ZNF460 | 0.715961 | 3 | 0 | 3 |
| PDE1B | TUBB4A | 0.701809 | 3 | 0 | 3 |
| PDE1B | TEX15 | 0.697207 | 3 | 0 | 3 |
| PDE1B | ARRB2 | 0.694822 | 4 | 0 | 3 |
| PDE1B | OR1A1 | 0.689535 | 4 | 0 | 4 |
| PDE1B | KRT76 | 0.688592 | 4 | 0 | 3 |
| PDE1B | SLC10A1 | 0.681162 | 4 | 0 | 4 |
| PDE1B | SIGLEC7 | 0.679816 | 8 | 0 | 7 |
| PDE1B | NXPE4 | 0.679663 | 4 | 0 | 4 |
| PDE1B | LINC00563 | 0.669494 | 3 | 0 | 3 |
| PDE1B | AP4S1 | 0.669479 | 5 | 0 | 4 |
| PDE1B | SNAI3 | 0.665669 | 3 | 0 | 3 |
| PDE1B | MYL2 | 0.665256 | 5 | 0 | 5 |
| PDE1B | MMP8 | 0.662341 | 3 | 0 | 3 |
| PDE1B | GRIN2A | 0.660609 | 5 | 0 | 5 |
| PDE1B | PCSK6 | 0.656749 | 6 | 0 | 4 |
| PDE1B | PNMA3 | 0.654972 | 7 | 0 | 6 |
| PDE1B | KIR2DS5 | 0.654908 | 7 | 0 | 5 |
| PDE1B | PAX8 | 0.654663 | 5 | 0 | 4 |
| PDE1B | KIF5A | 0.652919 | 4 | 0 | 4 |
For details and further investigation, click here