| Full name: phosphatidylethanolamine binding protein 4 | Alias Symbol: MGC22776|CORK1|hPEBP4 | ||
| Type: protein-coding gene | Cytoband: 8p21.3 | ||
| Entrez ID: 157310 | HGNC ID: HGNC:28319 | Ensembl Gene: ENSG00000134020 | OMIM ID: 612473 |
| Drug and gene relationship at DGIdb | |||
Expression of PEBP4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PEBP4 | 157310 | 227848_at | -0.4411 | 0.2966 | |
| GSE26886 | PEBP4 | 157310 | 227848_at | 0.2182 | 0.0723 | |
| GSE45670 | PEBP4 | 157310 | 227848_at | -0.2481 | 0.0287 | |
| GSE53622 | PEBP4 | 157310 | 41847 | -0.3700 | 0.0208 | |
| GSE53624 | PEBP4 | 157310 | 41847 | -0.2891 | 0.0556 | |
| GSE63941 | PEBP4 | 157310 | 227848_at | 0.0314 | 0.8642 | |
| GSE77861 | PEBP4 | 157310 | 227848_at | -0.1111 | 0.2537 | |
| GSE97050 | PEBP4 | 157310 | A_33_P3627001 | -0.3863 | 0.5137 | |
| TCGA | PEBP4 | 157310 | RNAseq | -4.3860 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
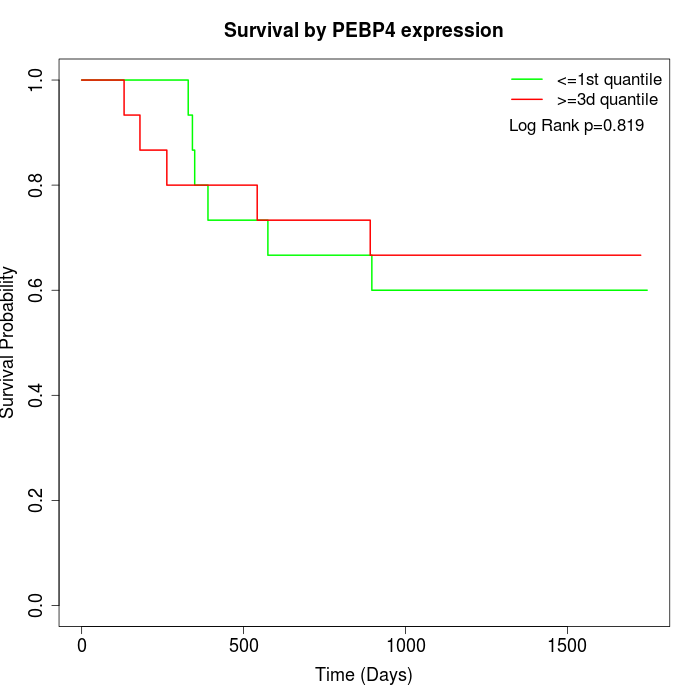
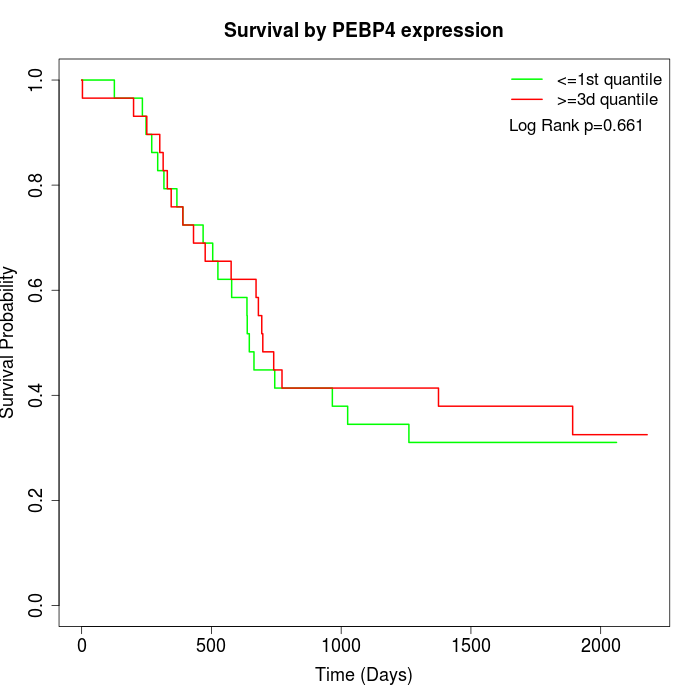
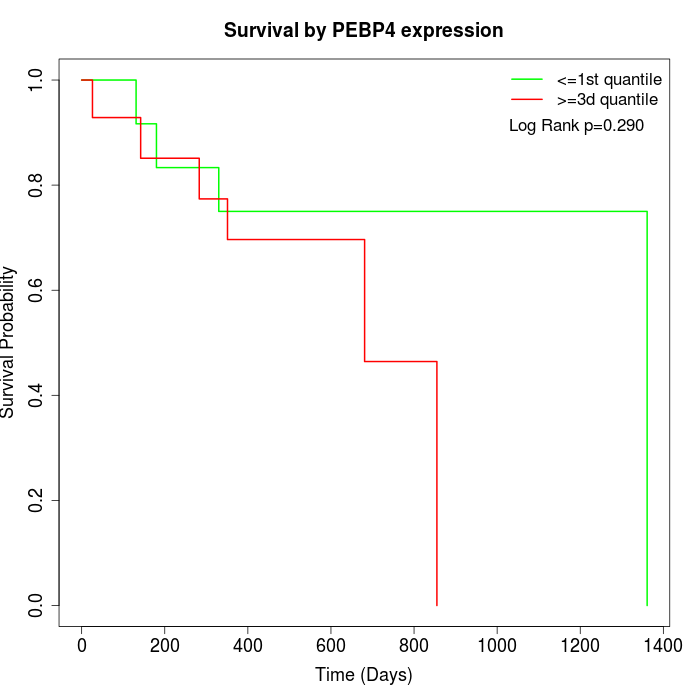
Survival by PEBP4 expression:
Note: Click image to view full size file.
Copy number change of PEBP4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PEBP4 | 157310 | 4 | 10 | 16 | |
| GSE20123 | PEBP4 | 157310 | 4 | 10 | 16 | |
| GSE43470 | PEBP4 | 157310 | 4 | 9 | 30 | |
| GSE46452 | PEBP4 | 157310 | 13 | 13 | 33 | |
| GSE47630 | PEBP4 | 157310 | 10 | 8 | 22 | |
| GSE54993 | PEBP4 | 157310 | 2 | 14 | 54 | |
| GSE54994 | PEBP4 | 157310 | 9 | 17 | 27 | |
| GSE60625 | PEBP4 | 157310 | 3 | 0 | 8 | |
| GSE74703 | PEBP4 | 157310 | 4 | 7 | 25 | |
| GSE74704 | PEBP4 | 157310 | 3 | 7 | 10 | |
| TCGA | PEBP4 | 157310 | 14 | 43 | 39 |
Total number of gains: 70; Total number of losses: 138; Total Number of normals: 280.
Somatic mutations of PEBP4:
Generating mutation plots.
Highly correlated genes for PEBP4:
Showing top 20/150 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PEBP4 | CORO6 | 0.765063 | 3 | 0 | 3 |
| PEBP4 | SLC35F1 | 0.749354 | 3 | 0 | 3 |
| PEBP4 | USP2 | 0.731761 | 3 | 0 | 3 |
| PEBP4 | PCDH9 | 0.715201 | 3 | 0 | 3 |
| PEBP4 | RYR2 | 0.708912 | 3 | 0 | 3 |
| PEBP4 | ANO5 | 0.702569 | 3 | 0 | 3 |
| PEBP4 | KCNN3 | 0.699815 | 4 | 0 | 3 |
| PEBP4 | NRXN3 | 0.698503 | 3 | 0 | 3 |
| PEBP4 | IQUB | 0.689894 | 3 | 0 | 3 |
| PEBP4 | PDE3A | 0.6865 | 4 | 0 | 4 |
| PEBP4 | CHD5 | 0.685347 | 3 | 0 | 3 |
| PEBP4 | TTLL7 | 0.676314 | 3 | 0 | 3 |
| PEBP4 | CCDC136 | 0.671125 | 4 | 0 | 3 |
| PEBP4 | MYL9 | 0.667583 | 3 | 0 | 3 |
| PEBP4 | CARTPT | 0.664359 | 3 | 0 | 3 |
| PEBP4 | HECTD2 | 0.663514 | 3 | 0 | 3 |
| PEBP4 | GABRA5 | 0.662808 | 4 | 0 | 4 |
| PEBP4 | YPEL1 | 0.657682 | 4 | 0 | 3 |
| PEBP4 | TMEM8B | 0.655938 | 4 | 0 | 3 |
| PEBP4 | PPP1R1A | 0.655627 | 5 | 0 | 4 |
For details and further investigation, click here