| Full name: phosphorylase kinase catalytic subunit gamma 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 16p11.2 | ||
| Entrez ID: 5261 | HGNC ID: HGNC:8931 | Ensembl Gene: ENSG00000156873 | OMIM ID: 172471 |
| Drug and gene relationship at DGIdb | |||
PHKG2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04020 | Calcium signaling pathway | |
| hsa04910 | Insulin signaling pathway | |
| hsa04922 | Glucagon signaling pathway |
Expression of PHKG2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PHKG2 | 5261 | 203709_at | 0.3779 | 0.2537 | |
| GSE20347 | PHKG2 | 5261 | 203709_at | 0.3359 | 0.0020 | |
| GSE23400 | PHKG2 | 5261 | 203709_at | 0.2597 | 0.0000 | |
| GSE26886 | PHKG2 | 5261 | 203709_at | 0.3959 | 0.0057 | |
| GSE29001 | PHKG2 | 5261 | 203709_at | 0.4338 | 0.0686 | |
| GSE38129 | PHKG2 | 5261 | 203709_at | 0.3360 | 0.0001 | |
| GSE45670 | PHKG2 | 5261 | 203709_at | 0.5293 | 0.0000 | |
| GSE53622 | PHKG2 | 5261 | 36781 | 0.5133 | 0.0000 | |
| GSE53624 | PHKG2 | 5261 | 36781 | 0.6707 | 0.0000 | |
| GSE63941 | PHKG2 | 5261 | 203709_at | 0.4886 | 0.1160 | |
| GSE77861 | PHKG2 | 5261 | 203709_at | 0.3868 | 0.1581 | |
| GSE97050 | PHKG2 | 5261 | A_24_P296457 | 0.3625 | 0.1939 | |
| SRP007169 | PHKG2 | 5261 | RNAseq | 0.0486 | 0.9176 | |
| SRP008496 | PHKG2 | 5261 | RNAseq | 0.4962 | 0.0883 | |
| SRP064894 | PHKG2 | 5261 | RNAseq | 0.3412 | 0.0110 | |
| SRP133303 | PHKG2 | 5261 | RNAseq | 0.4345 | 0.0034 | |
| SRP159526 | PHKG2 | 5261 | RNAseq | 0.5331 | 0.0998 | |
| SRP193095 | PHKG2 | 5261 | RNAseq | 0.3923 | 0.0006 | |
| SRP219564 | PHKG2 | 5261 | RNAseq | 0.4771 | 0.0736 | |
| TCGA | PHKG2 | 5261 | RNAseq | 0.0781 | 0.1659 |
Upregulated datasets: 0; Downregulated datasets: 0.
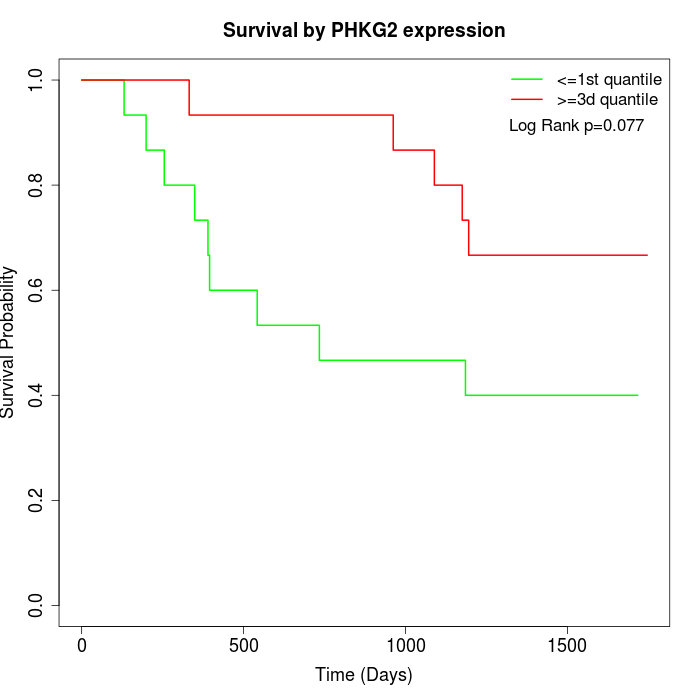
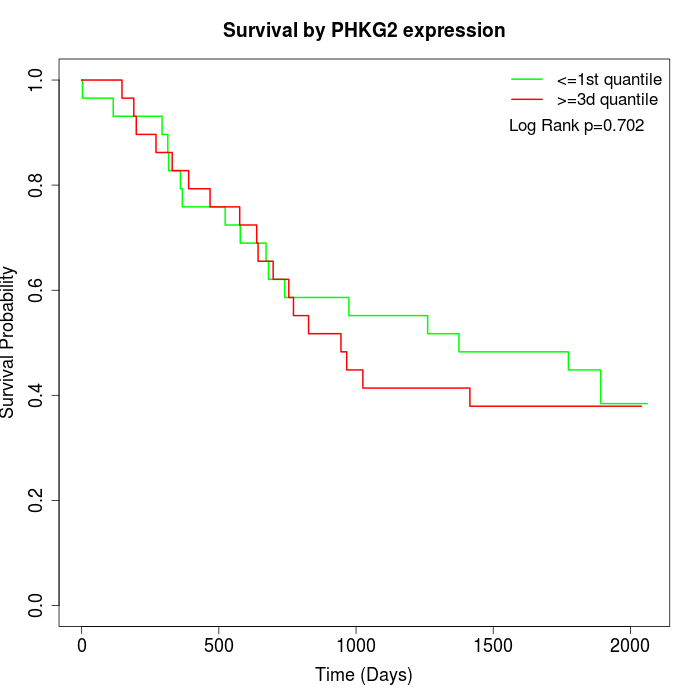
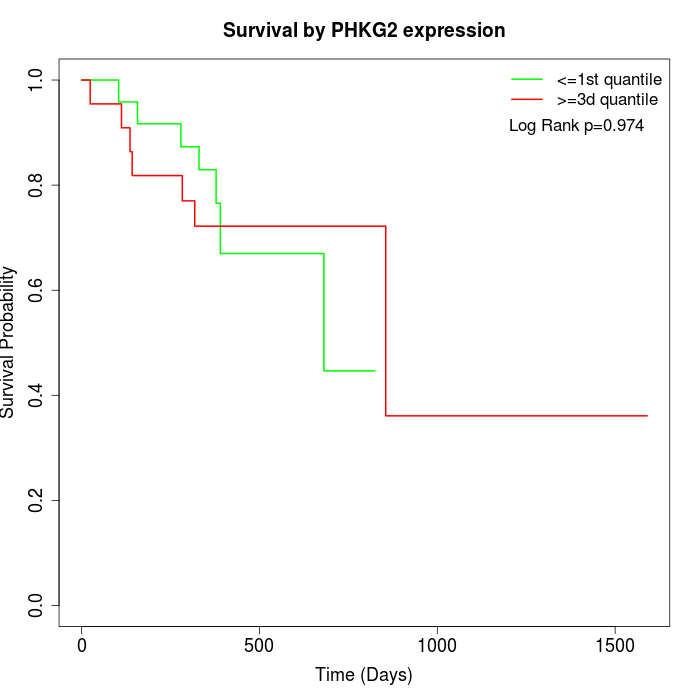
Survival by PHKG2 expression:
Note: Click image to view full size file.
Copy number change of PHKG2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PHKG2 | 5261 | 5 | 5 | 20 | |
| GSE20123 | PHKG2 | 5261 | 5 | 4 | 21 | |
| GSE43470 | PHKG2 | 5261 | 3 | 3 | 37 | |
| GSE46452 | PHKG2 | 5261 | 38 | 1 | 20 | |
| GSE47630 | PHKG2 | 5261 | 11 | 7 | 22 | |
| GSE54993 | PHKG2 | 5261 | 2 | 5 | 63 | |
| GSE54994 | PHKG2 | 5261 | 5 | 9 | 39 | |
| GSE60625 | PHKG2 | 5261 | 4 | 0 | 7 | |
| GSE74703 | PHKG2 | 5261 | 3 | 2 | 31 | |
| GSE74704 | PHKG2 | 5261 | 3 | 2 | 15 | |
| TCGA | PHKG2 | 5261 | 20 | 10 | 66 |
Total number of gains: 99; Total number of losses: 48; Total Number of normals: 341.
Somatic mutations of PHKG2:
Generating mutation plots.
Highly correlated genes for PHKG2:
Showing top 20/1379 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PHKG2 | INTS9 | 0.816537 | 3 | 0 | 3 |
| PHKG2 | SPNS1 | 0.787982 | 5 | 0 | 5 |
| PHKG2 | C7orf50 | 0.765424 | 3 | 0 | 3 |
| PHKG2 | KTI12 | 0.763645 | 3 | 0 | 3 |
| PHKG2 | TRAF7 | 0.738583 | 6 | 0 | 5 |
| PHKG2 | DNMBP | 0.731744 | 3 | 0 | 3 |
| PHKG2 | ITGB8 | 0.728062 | 3 | 0 | 3 |
| PHKG2 | FBXL19 | 0.72427 | 6 | 0 | 6 |
| PHKG2 | PRKACA | 0.723483 | 3 | 0 | 3 |
| PHKG2 | DTD1 | 0.722384 | 3 | 0 | 3 |
| PHKG2 | EHD1 | 0.719451 | 3 | 0 | 3 |
| PHKG2 | SMAD3 | 0.710763 | 3 | 0 | 3 |
| PHKG2 | SLC35B2 | 0.708755 | 3 | 0 | 3 |
| PHKG2 | ZNF417 | 0.707582 | 4 | 0 | 4 |
| PHKG2 | ANKRD11 | 0.704734 | 3 | 0 | 3 |
| PHKG2 | DNTTIP1 | 0.703175 | 7 | 0 | 6 |
| PHKG2 | NDUFB4 | 0.702706 | 3 | 0 | 3 |
| PHKG2 | OCEL1 | 0.700117 | 4 | 0 | 3 |
| PHKG2 | YWHAG | 0.698675 | 4 | 0 | 4 |
| PHKG2 | IRF2BPL | 0.697129 | 4 | 0 | 3 |
For details and further investigation, click here