| Full name: phosphatidylinositol binding clathrin assembly protein | Alias Symbol: CALM|CLTH | ||
| Type: protein-coding gene | Cytoband: 11q14.2 | ||
| Entrez ID: 8301 | HGNC ID: HGNC:15514 | Ensembl Gene: ENSG00000073921 | OMIM ID: 603025 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of PICALM:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PICALM | 8301 | 212506_at | -0.1121 | 0.7093 | |
| GSE20347 | PICALM | 8301 | 212506_at | -0.1839 | 0.2849 | |
| GSE23400 | PICALM | 8301 | 212506_at | -0.0485 | 0.4828 | |
| GSE26886 | PICALM | 8301 | 212506_at | -0.6532 | 0.0012 | |
| GSE29001 | PICALM | 8301 | 212506_at | -0.4234 | 0.1367 | |
| GSE38129 | PICALM | 8301 | 212506_at | -0.1832 | 0.0990 | |
| GSE45670 | PICALM | 8301 | 212506_at | -0.0534 | 0.6973 | |
| GSE53622 | PICALM | 8301 | 102163 | 0.0321 | 0.6389 | |
| GSE53624 | PICALM | 8301 | 102163 | -0.1226 | 0.0172 | |
| GSE63941 | PICALM | 8301 | 212506_at | -0.9526 | 0.0432 | |
| GSE77861 | PICALM | 8301 | 212506_at | -0.1877 | 0.4422 | |
| GSE97050 | PICALM | 8301 | A_23_P203658 | 0.5258 | 0.2494 | |
| SRP007169 | PICALM | 8301 | RNAseq | -0.9925 | 0.0102 | |
| SRP008496 | PICALM | 8301 | RNAseq | -0.8253 | 0.0003 | |
| SRP064894 | PICALM | 8301 | RNAseq | -0.1212 | 0.5833 | |
| SRP133303 | PICALM | 8301 | RNAseq | 0.0031 | 0.9854 | |
| SRP159526 | PICALM | 8301 | RNAseq | -0.5910 | 0.0312 | |
| SRP193095 | PICALM | 8301 | RNAseq | -0.5063 | 0.0004 | |
| SRP219564 | PICALM | 8301 | RNAseq | -0.3733 | 0.1899 | |
| TCGA | PICALM | 8301 | RNAseq | -0.0262 | 0.5231 |
Upregulated datasets: 0; Downregulated datasets: 0.
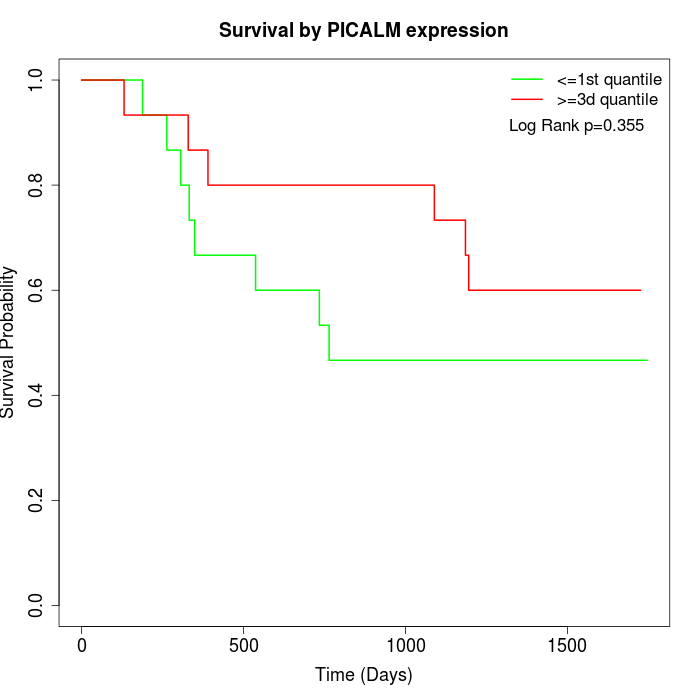
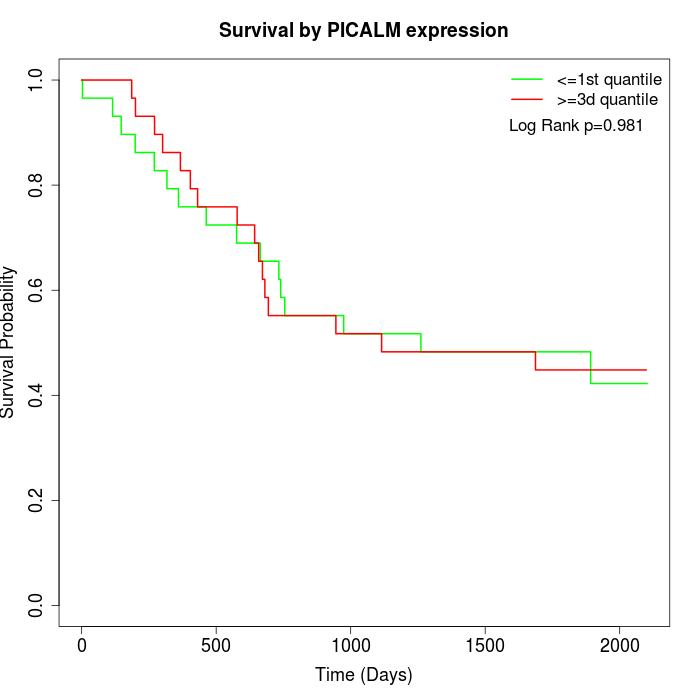
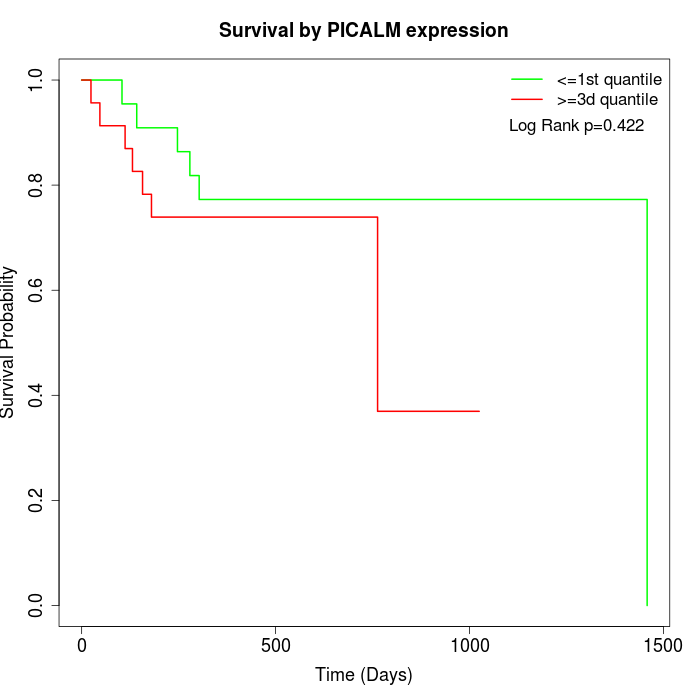
Survival by PICALM expression:
Note: Click image to view full size file.
Copy number change of PICALM:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PICALM | 8301 | 4 | 9 | 17 | |
| GSE20123 | PICALM | 8301 | 4 | 9 | 17 | |
| GSE43470 | PICALM | 8301 | 3 | 4 | 36 | |
| GSE46452 | PICALM | 8301 | 5 | 24 | 30 | |
| GSE47630 | PICALM | 8301 | 2 | 19 | 19 | |
| GSE54993 | PICALM | 8301 | 8 | 2 | 60 | |
| GSE54994 | PICALM | 8301 | 6 | 18 | 29 | |
| GSE60625 | PICALM | 8301 | 0 | 3 | 8 | |
| GSE74703 | PICALM | 8301 | 2 | 3 | 31 | |
| GSE74704 | PICALM | 8301 | 3 | 6 | 11 | |
| TCGA | PICALM | 8301 | 16 | 39 | 41 |
Total number of gains: 53; Total number of losses: 136; Total Number of normals: 299.
Somatic mutations of PICALM:
Generating mutation plots.
Highly correlated genes for PICALM:
Showing top 20/67 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PICALM | KLHL18 | 0.702258 | 3 | 0 | 3 |
| PICALM | HACE1 | 0.65659 | 3 | 0 | 3 |
| PICALM | SEC63 | 0.648547 | 3 | 0 | 3 |
| PICALM | SMIM15 | 0.648197 | 3 | 0 | 3 |
| PICALM | PPIC | 0.629393 | 3 | 0 | 3 |
| PICALM | IL10RB | 0.628011 | 3 | 0 | 3 |
| PICALM | SMIM14 | 0.621031 | 4 | 0 | 3 |
| PICALM | SMPD1 | 0.61777 | 3 | 0 | 3 |
| PICALM | PLAA | 0.617724 | 4 | 0 | 4 |
| PICALM | KMT2A | 0.606978 | 3 | 0 | 3 |
| PICALM | LPIN1 | 0.605429 | 4 | 0 | 3 |
| PICALM | VSIG10L | 0.60457 | 3 | 0 | 3 |
| PICALM | SMPD2 | 0.599226 | 3 | 0 | 3 |
| PICALM | LACC1 | 0.596068 | 4 | 0 | 3 |
| PICALM | SLC25A51 | 0.590723 | 3 | 0 | 3 |
| PICALM | PRMT9 | 0.589976 | 3 | 0 | 3 |
| PICALM | STT3A | 0.587753 | 4 | 0 | 3 |
| PICALM | CNDP2 | 0.585336 | 3 | 0 | 3 |
| PICALM | SNX16 | 0.583159 | 4 | 0 | 3 |
| PICALM | CLPX | 0.581149 | 5 | 0 | 4 |
For details and further investigation, click here