| Full name: phosphatidylinositol glycan anchor biosynthesis class V | Alias Symbol: FLJ20477 | ||
| Type: protein-coding gene | Cytoband: 1p36.11 | ||
| Entrez ID: 55650 | HGNC ID: HGNC:26031 | Ensembl Gene: ENSG00000060642 | OMIM ID: 610274 |
| Drug and gene relationship at DGIdb | |||
Expression of PIGV:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PIGV | 55650 | 219238_at | -0.2943 | 0.6554 | |
| GSE20347 | PIGV | 55650 | 219238_at | -0.2512 | 0.0778 | |
| GSE23400 | PIGV | 55650 | 219238_at | 0.2932 | 0.0000 | |
| GSE26886 | PIGV | 55650 | 219238_at | -0.2009 | 0.4059 | |
| GSE29001 | PIGV | 55650 | 219238_at | 0.1435 | 0.5500 | |
| GSE38129 | PIGV | 55650 | 219238_at | -0.0298 | 0.8197 | |
| GSE45670 | PIGV | 55650 | 219238_at | 0.1731 | 0.2307 | |
| GSE53622 | PIGV | 55650 | 14932 | -0.0363 | 0.7431 | |
| GSE53624 | PIGV | 55650 | 14932 | -0.0223 | 0.8544 | |
| GSE63941 | PIGV | 55650 | 219238_at | -0.0893 | 0.8218 | |
| GSE77861 | PIGV | 55650 | 219238_at | -0.0867 | 0.7699 | |
| GSE97050 | PIGV | 55650 | A_23_P46378 | 0.0121 | 0.9829 | |
| SRP007169 | PIGV | 55650 | RNAseq | -0.4589 | 0.4210 | |
| SRP008496 | PIGV | 55650 | RNAseq | -0.4577 | 0.1332 | |
| SRP064894 | PIGV | 55650 | RNAseq | 0.2594 | 0.0940 | |
| SRP133303 | PIGV | 55650 | RNAseq | 0.0761 | 0.7142 | |
| SRP159526 | PIGV | 55650 | RNAseq | -0.1856 | 0.4689 | |
| SRP193095 | PIGV | 55650 | RNAseq | -0.3159 | 0.0131 | |
| SRP219564 | PIGV | 55650 | RNAseq | 0.1312 | 0.7169 | |
| TCGA | PIGV | 55650 | RNAseq | 0.0014 | 0.9814 |
Upregulated datasets: 0; Downregulated datasets: 0.
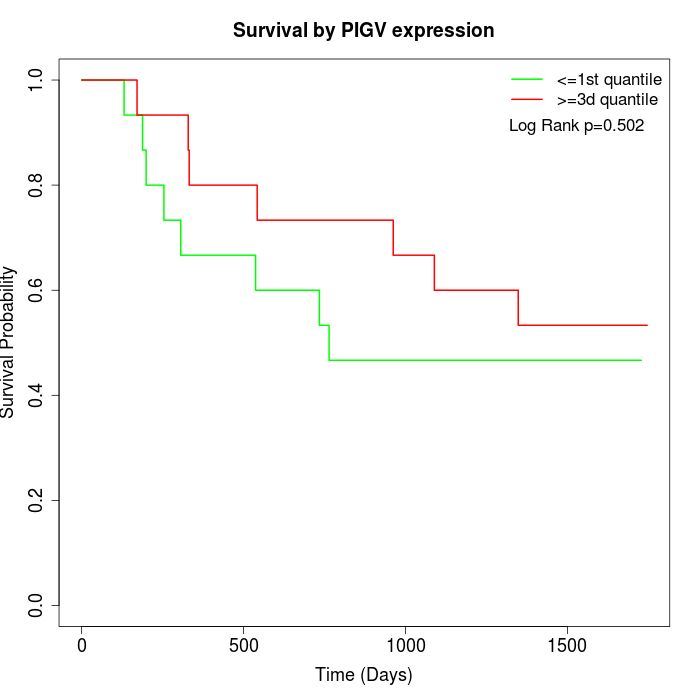
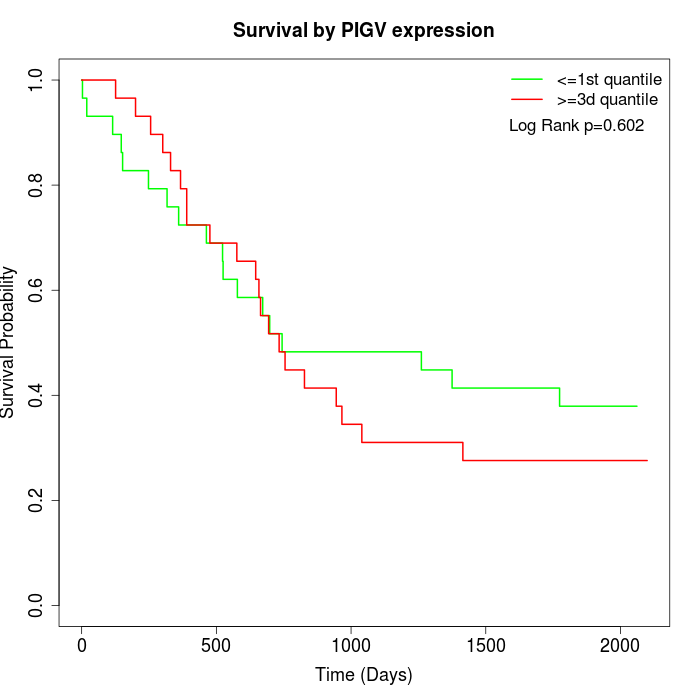
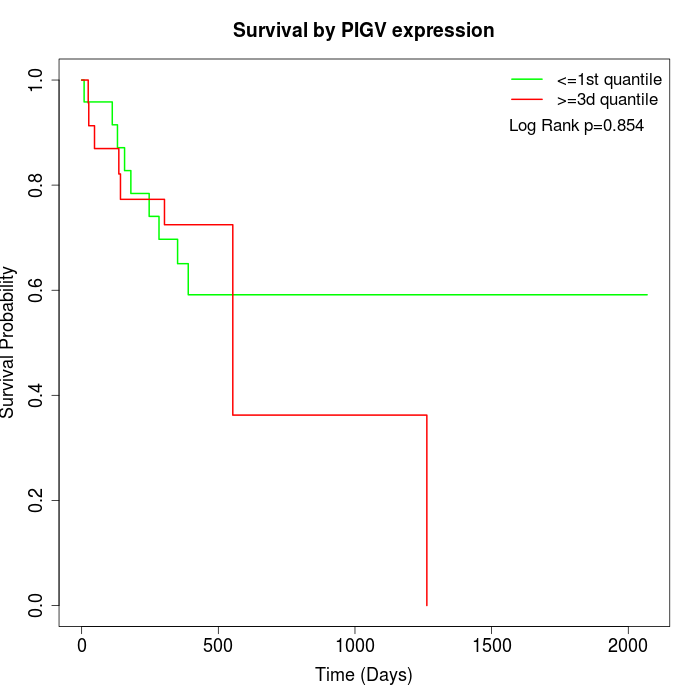
Survival by PIGV expression:
Note: Click image to view full size file.
Copy number change of PIGV:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PIGV | 55650 | 0 | 6 | 24 | |
| GSE20123 | PIGV | 55650 | 0 | 5 | 25 | |
| GSE43470 | PIGV | 55650 | 2 | 6 | 35 | |
| GSE46452 | PIGV | 55650 | 5 | 1 | 53 | |
| GSE47630 | PIGV | 55650 | 8 | 3 | 29 | |
| GSE54993 | PIGV | 55650 | 2 | 1 | 67 | |
| GSE54994 | PIGV | 55650 | 10 | 4 | 39 | |
| GSE60625 | PIGV | 55650 | 0 | 0 | 11 | |
| GSE74703 | PIGV | 55650 | 1 | 4 | 31 | |
| GSE74704 | PIGV | 55650 | 0 | 0 | 20 | |
| TCGA | PIGV | 55650 | 9 | 25 | 62 |
Total number of gains: 37; Total number of losses: 55; Total Number of normals: 396.
Somatic mutations of PIGV:
Generating mutation plots.
Highly correlated genes for PIGV:
Showing top 20/251 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PIGV | GNG12 | 0.768531 | 3 | 0 | 3 |
| PIGV | LEPROT | 0.745609 | 3 | 0 | 3 |
| PIGV | MBD1 | 0.712478 | 3 | 0 | 3 |
| PIGV | SLFN12 | 0.708002 | 3 | 0 | 3 |
| PIGV | BAG5 | 0.701053 | 3 | 0 | 3 |
| PIGV | FAM162B | 0.70072 | 3 | 0 | 3 |
| PIGV | GTF2B | 0.698475 | 4 | 0 | 3 |
| PIGV | AIMP2 | 0.695546 | 3 | 0 | 3 |
| PIGV | GPSM2 | 0.686643 | 3 | 0 | 3 |
| PIGV | NDUFA9 | 0.684049 | 3 | 0 | 3 |
| PIGV | USP40 | 0.676502 | 3 | 0 | 3 |
| PIGV | ANKMY2 | 0.675758 | 3 | 0 | 3 |
| PIGV | SAP130 | 0.675687 | 3 | 0 | 3 |
| PIGV | MLH1 | 0.674427 | 4 | 0 | 4 |
| PIGV | LYSMD3 | 0.673346 | 4 | 0 | 3 |
| PIGV | ZSWIM5 | 0.672204 | 4 | 0 | 4 |
| PIGV | XPO4 | 0.670682 | 4 | 0 | 3 |
| PIGV | ETS2 | 0.670621 | 3 | 0 | 3 |
| PIGV | ELP3 | 0.669836 | 3 | 0 | 3 |
| PIGV | MAN1A2 | 0.667229 | 3 | 0 | 3 |
For details and further investigation, click here