| Full name: phosphatidylinositol 3-kinase catalytic subunit type 3 | Alias Symbol: Vps34 | ||
| Type: protein-coding gene | Cytoband: 18q12.3 | ||
| Entrez ID: 5289 | HGNC ID: HGNC:8974 | Ensembl Gene: ENSG00000078142 | OMIM ID: 602609 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
PIK3C3 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05152 | Tuberculosis |
Expression of PIK3C3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PIK3C3 | 5289 | 204297_at | -0.2101 | 0.6144 | |
| GSE20347 | PIK3C3 | 5289 | 204297_at | -0.0947 | 0.5221 | |
| GSE23400 | PIK3C3 | 5289 | 204297_at | 0.0730 | 0.2139 | |
| GSE26886 | PIK3C3 | 5289 | 204297_at | -0.5131 | 0.0035 | |
| GSE29001 | PIK3C3 | 5289 | 204297_at | -0.0650 | 0.8352 | |
| GSE38129 | PIK3C3 | 5289 | 204297_at | -0.0437 | 0.7446 | |
| GSE45670 | PIK3C3 | 5289 | 204297_at | -0.0100 | 0.9615 | |
| GSE53622 | PIK3C3 | 5289 | 23919 | -0.4017 | 0.0000 | |
| GSE53624 | PIK3C3 | 5289 | 23919 | -0.1700 | 0.0181 | |
| GSE63941 | PIK3C3 | 5289 | 204297_at | -0.9586 | 0.0551 | |
| GSE77861 | PIK3C3 | 5289 | 204297_at | -0.0654 | 0.9112 | |
| GSE97050 | PIK3C3 | 5289 | A_23_P164536 | -0.1482 | 0.4368 | |
| SRP007169 | PIK3C3 | 5289 | RNAseq | -0.6500 | 0.0524 | |
| SRP008496 | PIK3C3 | 5289 | RNAseq | -0.1741 | 0.6064 | |
| SRP064894 | PIK3C3 | 5289 | RNAseq | -0.1826 | 0.1907 | |
| SRP133303 | PIK3C3 | 5289 | RNAseq | -0.2658 | 0.0339 | |
| SRP159526 | PIK3C3 | 5289 | RNAseq | -0.3462 | 0.1653 | |
| SRP193095 | PIK3C3 | 5289 | RNAseq | -0.2039 | 0.0198 | |
| SRP219564 | PIK3C3 | 5289 | RNAseq | -0.5119 | 0.0996 | |
| TCGA | PIK3C3 | 5289 | RNAseq | -0.0556 | 0.3174 |
Upregulated datasets: 0; Downregulated datasets: 0.
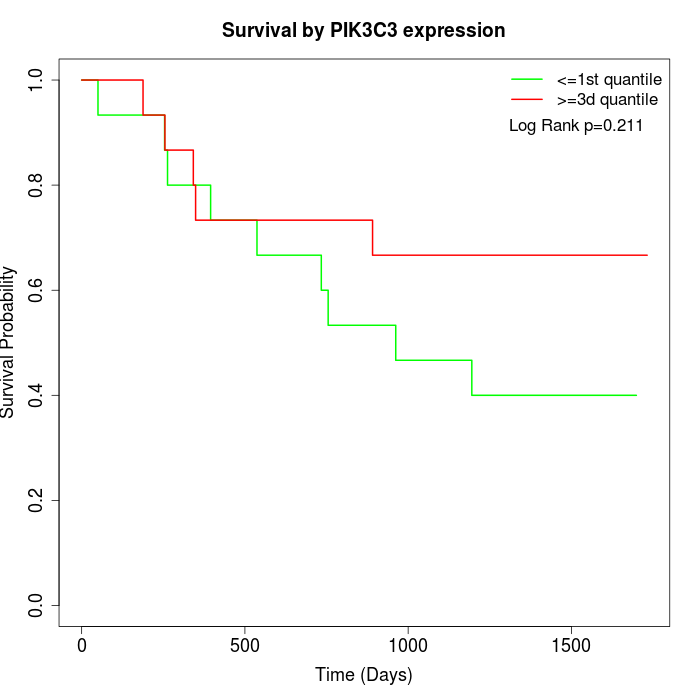
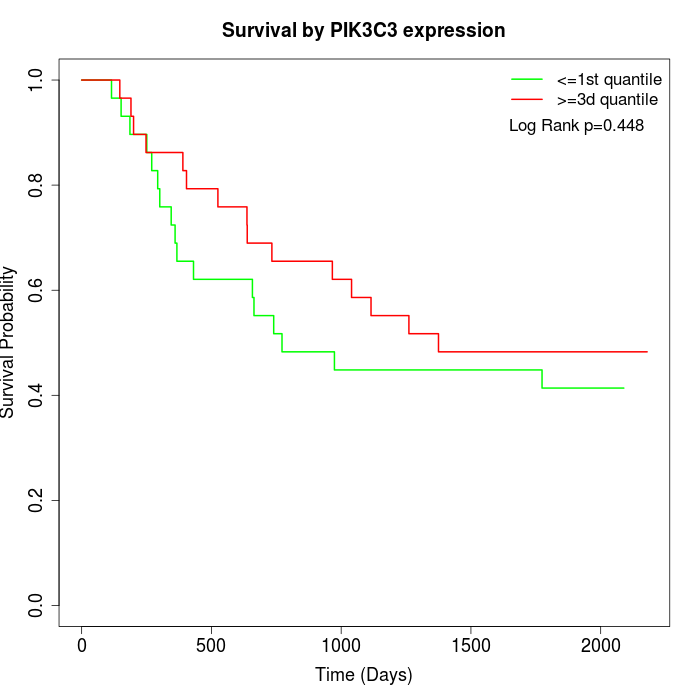
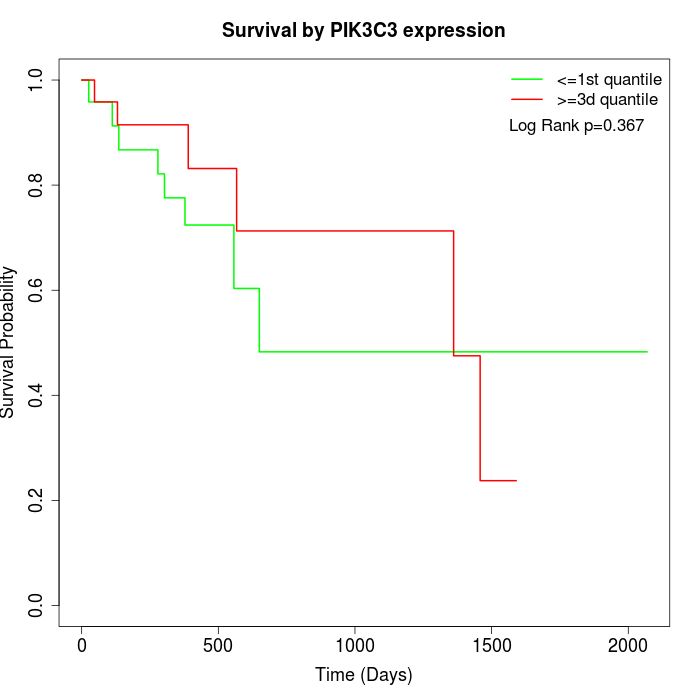
Survival by PIK3C3 expression:
Note: Click image to view full size file.
Copy number change of PIK3C3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PIK3C3 | 5289 | 1 | 6 | 23 | |
| GSE20123 | PIK3C3 | 5289 | 1 | 6 | 23 | |
| GSE43470 | PIK3C3 | 5289 | 0 | 3 | 40 | |
| GSE46452 | PIK3C3 | 5289 | 1 | 25 | 33 | |
| GSE47630 | PIK3C3 | 5289 | 5 | 20 | 15 | |
| GSE54993 | PIK3C3 | 5289 | 8 | 1 | 61 | |
| GSE54994 | PIK3C3 | 5289 | 2 | 15 | 36 | |
| GSE60625 | PIK3C3 | 5289 | 0 | 4 | 7 | |
| GSE74703 | PIK3C3 | 5289 | 0 | 3 | 33 | |
| GSE74704 | PIK3C3 | 5289 | 0 | 4 | 16 | |
| TCGA | PIK3C3 | 5289 | 14 | 39 | 43 |
Total number of gains: 32; Total number of losses: 126; Total Number of normals: 330.
Somatic mutations of PIK3C3:
Generating mutation plots.
Highly correlated genes for PIK3C3:
Showing top 20/613 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PIK3C3 | ACAP2 | 0.78391 | 3 | 0 | 3 |
| PIK3C3 | ZNF322 | 0.77885 | 3 | 0 | 3 |
| PIK3C3 | SLC35F5 | 0.773122 | 3 | 0 | 3 |
| PIK3C3 | BBS7 | 0.765734 | 3 | 0 | 3 |
| PIK3C3 | TET2 | 0.763021 | 3 | 0 | 3 |
| PIK3C3 | HOOK3 | 0.754878 | 3 | 0 | 3 |
| PIK3C3 | MFN1 | 0.750915 | 3 | 0 | 3 |
| PIK3C3 | TIMM21 | 0.743673 | 3 | 0 | 3 |
| PIK3C3 | NDUFS1 | 0.727606 | 5 | 0 | 5 |
| PIK3C3 | ORC4 | 0.72701 | 4 | 0 | 3 |
| PIK3C3 | HDHD2 | 0.723562 | 6 | 0 | 6 |
| PIK3C3 | RASA2 | 0.719266 | 3 | 0 | 3 |
| PIK3C3 | MTO1 | 0.713037 | 4 | 0 | 4 |
| PIK3C3 | KRAS | 0.70869 | 3 | 0 | 3 |
| PIK3C3 | RNF125 | 0.705583 | 4 | 0 | 4 |
| PIK3C3 | ZNF394 | 0.698384 | 3 | 0 | 3 |
| PIK3C3 | PPP4R2 | 0.696002 | 4 | 0 | 3 |
| PIK3C3 | ZNF136 | 0.69002 | 3 | 0 | 3 |
| PIK3C3 | ZNF649 | 0.689574 | 3 | 0 | 3 |
| PIK3C3 | LPIN2 | 0.688613 | 4 | 0 | 3 |
For details and further investigation, click here