| Full name: KRAS proto-oncogene, GTPase | Alias Symbol: KRAS1 | ||
| Type: protein-coding gene | Cytoband: 12p12.1 | ||
| Entrez ID: 3845 | HGNC ID: HGNC:6407 | Ensembl Gene: ENSG00000133703 | OMIM ID: 190070 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
KRAS involved pathways:
Expression of KRAS:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KRAS | 3845 | 204009_s_at | -0.1116 | 0.8563 | |
| GSE20347 | KRAS | 3845 | 214352_s_at | -0.2528 | 0.2578 | |
| GSE23400 | KRAS | 3845 | 204009_s_at | -0.0730 | 0.4859 | |
| GSE26886 | KRAS | 3845 | 204009_s_at | -0.7191 | 0.0030 | |
| GSE29001 | KRAS | 3845 | 204009_s_at | -0.1544 | 0.4241 | |
| GSE38129 | KRAS | 3845 | 214352_s_at | -0.2236 | 0.5252 | |
| GSE45670 | KRAS | 3845 | 204009_s_at | -0.0469 | 0.8183 | |
| GSE53622 | KRAS | 3845 | 148548 | -0.5233 | 0.0000 | |
| GSE53624 | KRAS | 3845 | 148548 | -0.6472 | 0.0000 | |
| GSE63941 | KRAS | 3845 | 204009_s_at | 0.0524 | 0.9334 | |
| GSE77861 | KRAS | 3845 | 204009_s_at | 0.0102 | 0.9705 | |
| GSE97050 | KRAS | 3845 | A_33_P3317815 | -0.4306 | 0.1581 | |
| SRP007169 | KRAS | 3845 | RNAseq | -0.7009 | 0.0288 | |
| SRP008496 | KRAS | 3845 | RNAseq | -0.3943 | 0.0605 | |
| SRP064894 | KRAS | 3845 | RNAseq | -0.6533 | 0.0069 | |
| SRP133303 | KRAS | 3845 | RNAseq | 0.2237 | 0.1573 | |
| SRP159526 | KRAS | 3845 | RNAseq | -0.4376 | 0.0647 | |
| SRP193095 | KRAS | 3845 | RNAseq | -0.5363 | 0.0000 | |
| SRP219564 | KRAS | 3845 | RNAseq | -0.7799 | 0.0165 | |
| TCGA | KRAS | 3845 | RNAseq | -0.0708 | 0.2039 |
Upregulated datasets: 0; Downregulated datasets: 0.
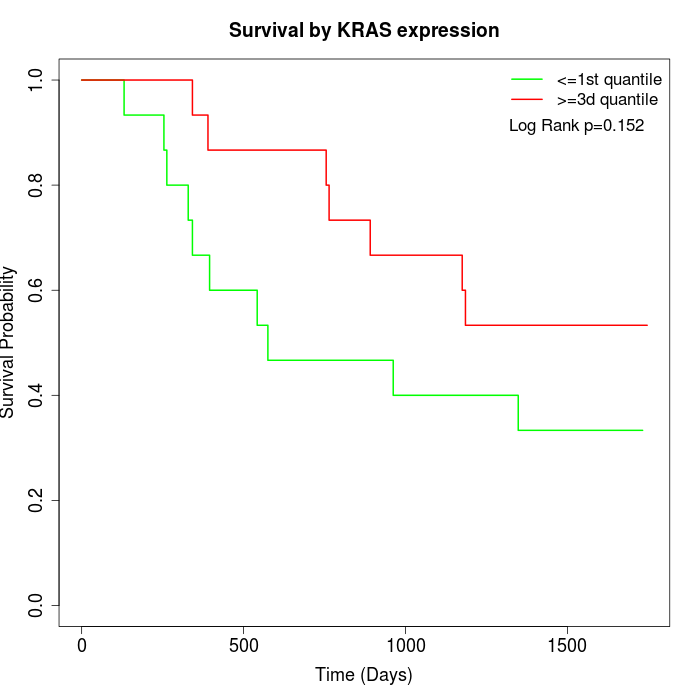
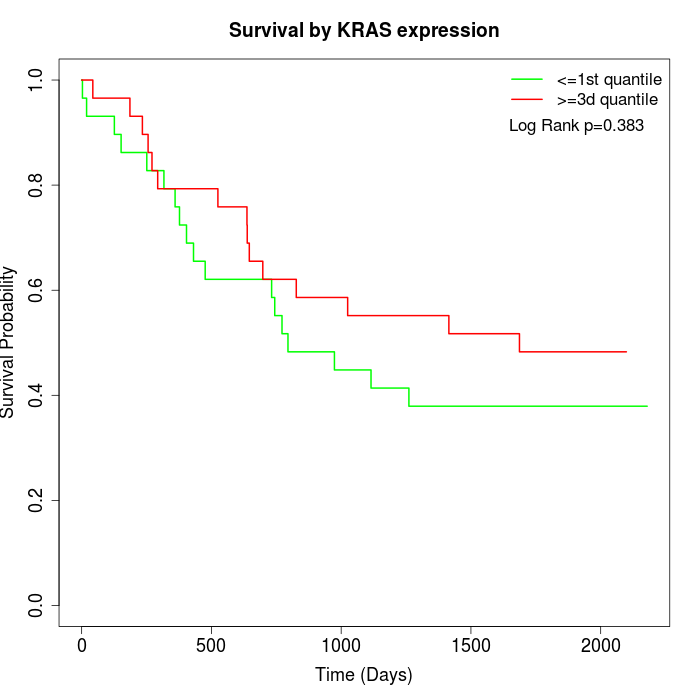
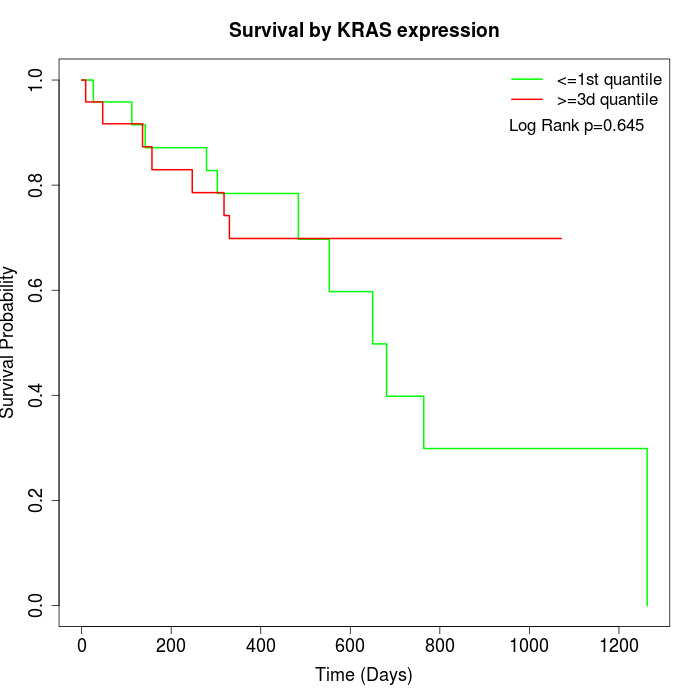
Survival by KRAS expression:
Note: Click image to view full size file.
Copy number change of KRAS:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KRAS | 3845 | 10 | 1 | 19 | |
| GSE20123 | KRAS | 3845 | 10 | 1 | 19 | |
| GSE43470 | KRAS | 3845 | 9 | 4 | 30 | |
| GSE46452 | KRAS | 3845 | 10 | 1 | 48 | |
| GSE47630 | KRAS | 3845 | 15 | 0 | 25 | |
| GSE54993 | KRAS | 3845 | 0 | 9 | 61 | |
| GSE54994 | KRAS | 3845 | 11 | 2 | 40 | |
| GSE60625 | KRAS | 3845 | 0 | 1 | 10 | |
| GSE74703 | KRAS | 3845 | 9 | 3 | 24 | |
| GSE74704 | KRAS | 3845 | 6 | 1 | 13 | |
| TCGA | KRAS | 3845 | 43 | 4 | 49 |
Total number of gains: 123; Total number of losses: 27; Total Number of normals: 338.
Somatic mutations of KRAS:
Generating mutation plots.
Highly correlated genes for KRAS:
Showing top 20/691 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KRAS | UBE2NL | 0.758453 | 3 | 0 | 3 |
| KRAS | GGA3 | 0.752025 | 3 | 0 | 3 |
| KRAS | CHD9 | 0.747546 | 3 | 0 | 3 |
| KRAS | LETMD1 | 0.740568 | 3 | 0 | 3 |
| KRAS | SENP8 | 0.7305 | 3 | 0 | 3 |
| KRAS | CBLL1 | 0.730087 | 3 | 0 | 3 |
| KRAS | EXOC4 | 0.725709 | 3 | 0 | 3 |
| KRAS | INSIG1 | 0.715505 | 3 | 0 | 3 |
| KRAS | ING4 | 0.712718 | 4 | 0 | 3 |
| KRAS | EPG5 | 0.712315 | 3 | 0 | 3 |
| KRAS | PIK3C3 | 0.70869 | 3 | 0 | 3 |
| KRAS | SREBF2 | 0.706521 | 3 | 0 | 3 |
| KRAS | ARL5B | 0.705643 | 3 | 0 | 3 |
| KRAS | KCTD15 | 0.705266 | 3 | 0 | 3 |
| KRAS | PRPF38A | 0.700359 | 3 | 0 | 3 |
| KRAS | CWC25 | 0.698776 | 3 | 0 | 3 |
| KRAS | CS | 0.697475 | 4 | 0 | 4 |
| KRAS | CTTNBP2NL | 0.693627 | 5 | 0 | 5 |
| KRAS | CPEB4 | 0.692647 | 5 | 0 | 5 |
| KRAS | COX5A | 0.692602 | 4 | 0 | 3 |
For details and further investigation, click here